Search Count: 100
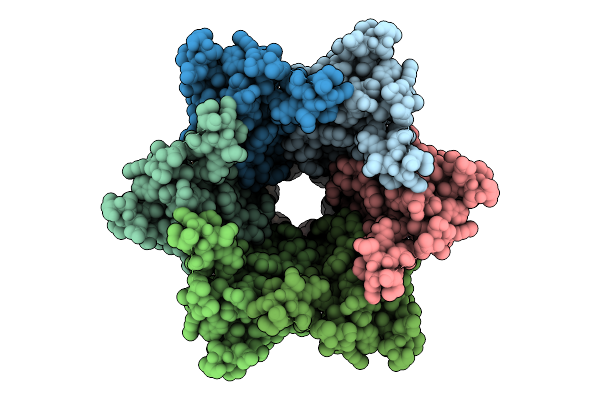 |
Connexin-32 (Cx32) Gap Junction Channel In Popc-Containing Nanodiscs In The Absence Of Chs
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
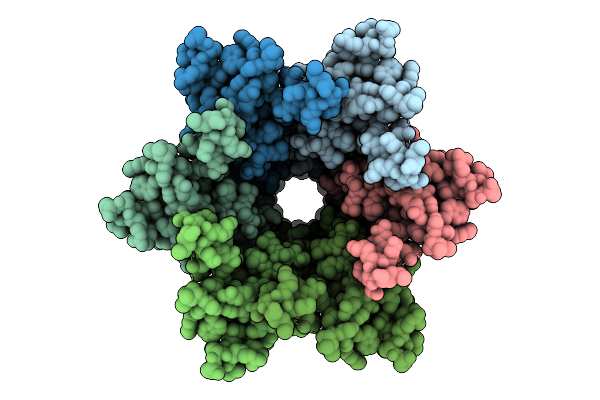 |
Connexin-32 (Cx32) W3S Mutant Gap Junction Channel In Popc-Containing Msp2N2 Nanodiscs
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
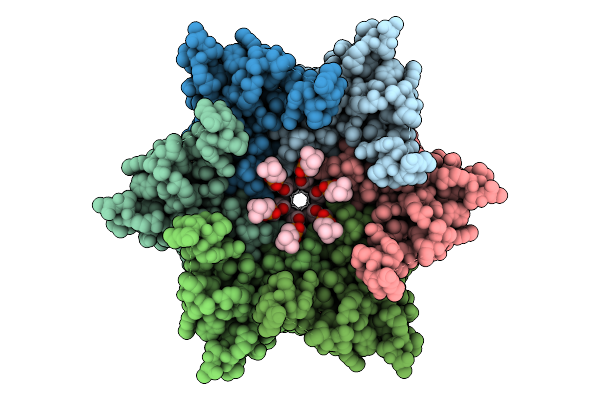 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: POV, CLR |
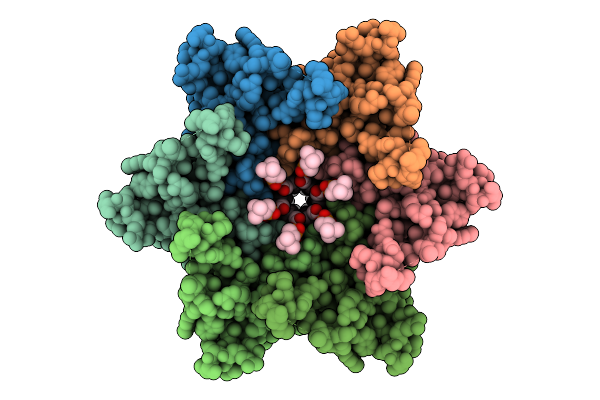 |
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: POV, CLR |
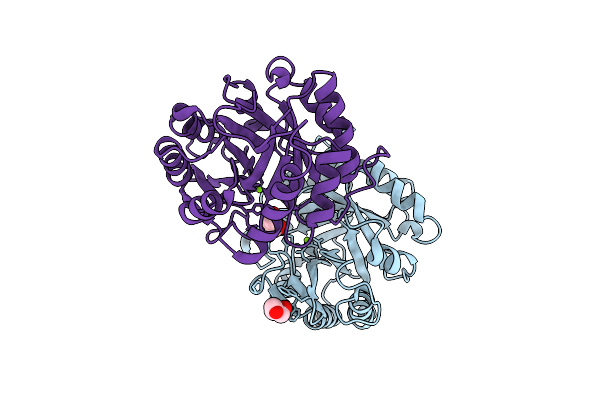 |
Crystal Structure Of Phospholipase D (Pld) From Arcanobacterium Haemolyticum
Organism: Arcanobacterium haemolyticum
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: HYDROLASE Ligands: GOL, MG |
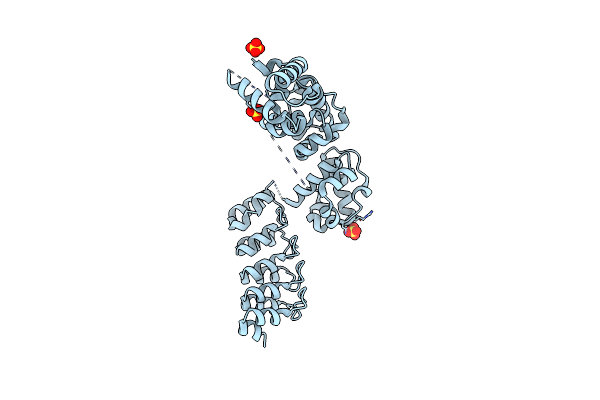 |
A Darpin Fused To The 3Tel Crystallization Chaperone Via A Gly-Gly-Gly Fusion
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.58 Å Release Date: 2024-12-04 Classification: PROTEIN BINDING Ligands: SO4 |
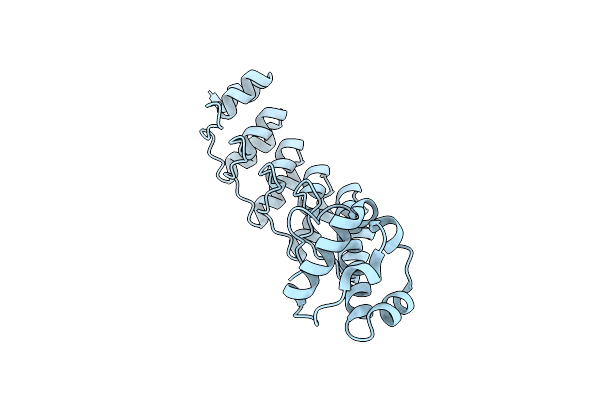 |
Crystal Structure Of A Darpin Fused To The 1Tel Crystallization Chaperone Via A Direct Helical Fusion In A 2-Fold Crystal Form
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-10-23 Classification: PROTEIN BINDING |
 |
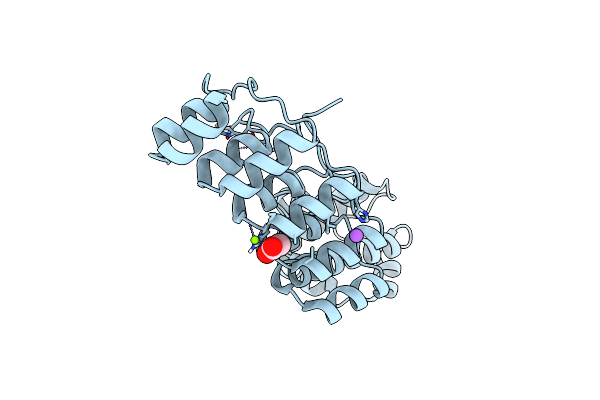
A Darpin Fused To The 1Tel Crystallization Chaperone Via A Helical Lys-Gln-Arg Linker
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2024-10-16 Classification: PROTEIN BINDING Ligands: HOH |
 |
Crystal Structure Of A Darpin Fused To The 1Tel Crystallization Chaperone Via A Direct Helical Fusion In A 3-Fold Crystal Form
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-10-09 Classification: PROTEIN BINDING Ligands: ACY, CL, MG, NA |
 |
A Darpin Fused To The 1Tel Crystallization Chaperone Via A Proline-Alanine Linker
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2024-10-09 Classification: PROTEIN BINDING Ligands: ACY, NA, CL |
 |
Crystal Structure Of Mutant Exfoliative Toxin C (Exhc) From Mammaliicoccus Sciuri
Organism: Mammaliicoccus sciuri
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-11-08 Classification: TOXIN |
 |
Crystal Structure Of Native Exfoliative Toxin C (Exhc) From Mammaliicoccus Sciuri
Organism: Mammaliicoccus sciuri
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2023-11-08 Classification: TOXIN |
 |
New Insights Into The P186 Flip And Oligomeric State Of Staphylococcus Aureus Exfoliative Toxin E: Implications For The Exfoliative Mechanism
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2022-11-02 Classification: TOXIN |
 |
Crystal Structure Of The O-Fucosylated Form Of Tsrs1-3 From The Human Thrombospondin 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-05-25 Classification: CELL ADHESION Ligands: FUC, EDO |
 |
Crystal Structure Of The Iron/Manganese Cambialistic Superoxide Dismutase From Rhodobacter Capsulatus Complex With Fe
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-12-01 Classification: OXIDOREDUCTASE Ligands: FE, CA |
 |
Crystal Structure Of The Iron/Manganese Cambialistic Superoxide Dismutase From Rhodobacter Capsulatus Complex With Mn
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-12-01 Classification: OXIDOREDUCTASE Ligands: MN, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2021-01-20 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-01-20 Classification: STRUCTURAL PROTEIN Ligands: SO4 |
 |
Organism: Acinetobacter sp. ver3
Method: X-RAY DIFFRACTION Release Date: 2020-07-22 Classification: OXIDOREDUCTASE Ligands: NAP, HEM, CL, UNX |
 |
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-05-20 Classification: SUGAR BINDING PROTEIN Ligands: GOL |

