Search Count: 108
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA-RNA HYBRID |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-10-15 Classification: DNA-RNA HYBRID |
 |
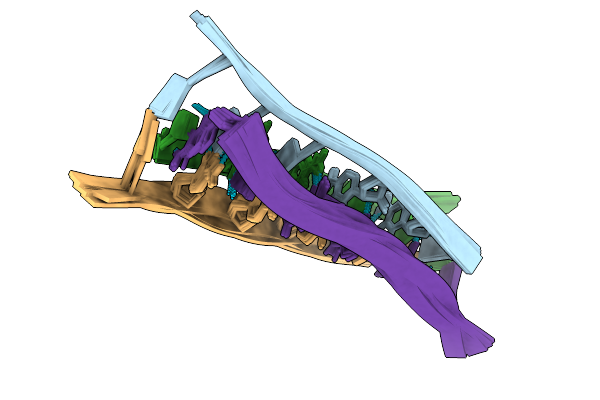
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: TRANSPORT PROTEIN Ligands: 377, GLU, CYZ |
 |
Organism: Rattus norvegicus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TRANSPORT PROTEIN Ligands: GLU, CYZ |
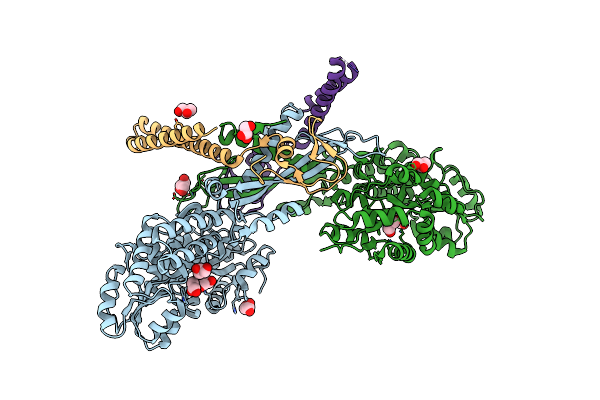 |
Organism: Escherichia coli k-12, Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-02-12 Classification: ANTITOXIN Ligands: GOL |
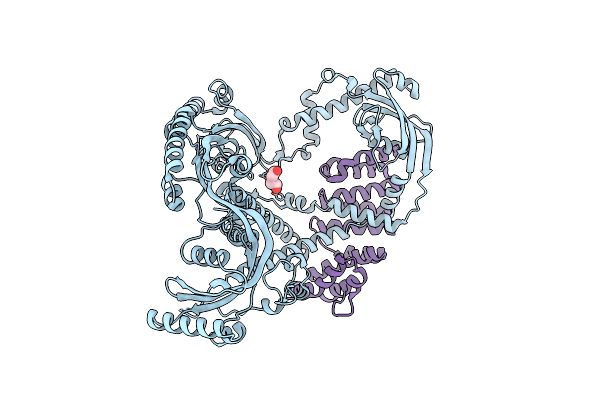 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-02-12 Classification: CHAPERONE Ligands: PEG |
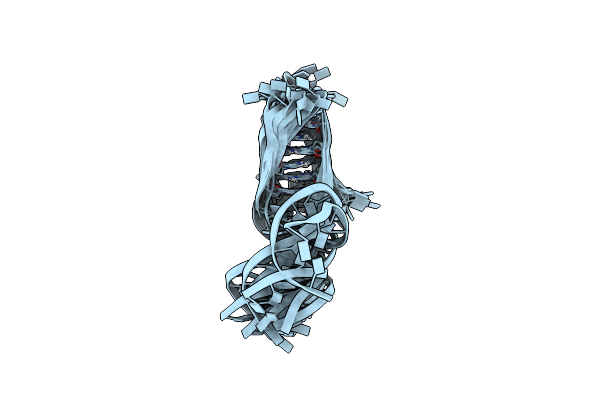 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2024-12-18 Classification: DNA BINDING PROTEIN |
 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
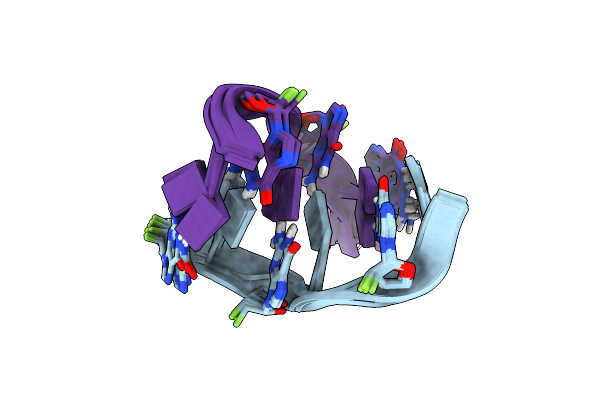 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
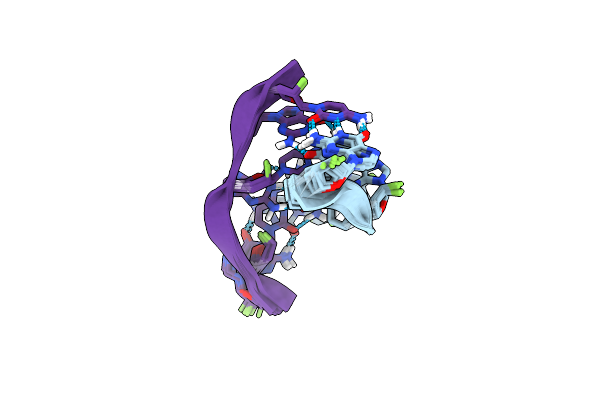 |
Formation Of Left-Handed Helices By C2'-Fluorinated Nucleic Acids Under Physiological Salt Conditions
|
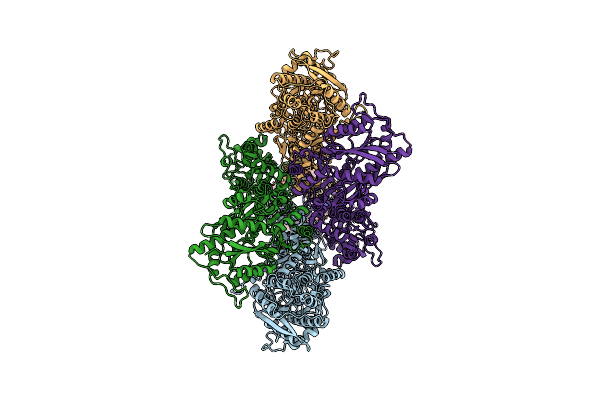 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: A1AB5 |
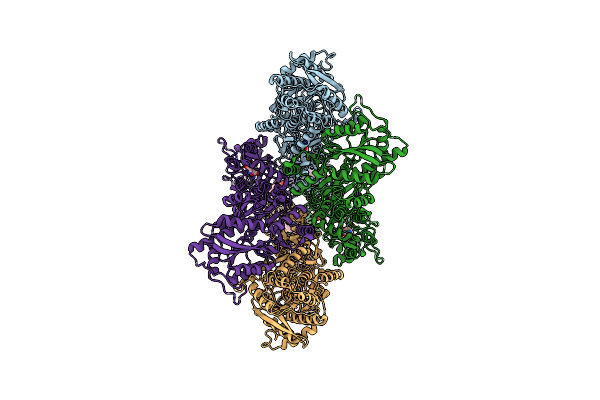 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: MEMBRANE PROTEIN/INHIBITOR Ligands: GLU, A1AB5 |
 |
Structure Of An I-Motif Domain With The Cytosine Analog 1,3-Diaza-2-Oxophenoxacione (Tc) At Neutral Ph
|
 |
 |
Organism: Cutibacterium acnes hl110pa3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-13 Classification: LYASE Ligands: GOL, PO4 |
 |
Organism: Cutibacterium acnes hl043pa1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-12-13 Classification: LYASE Ligands: EDO, GOL |
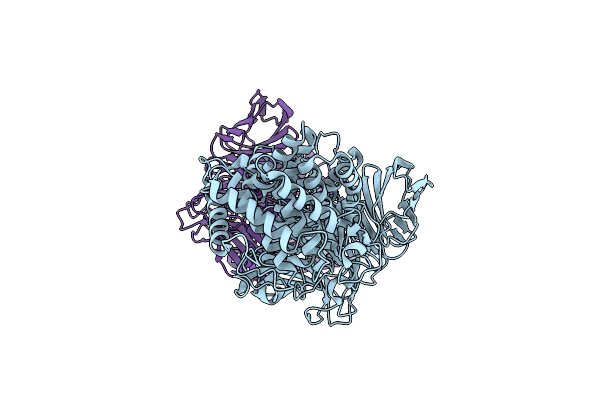 |
Crystal Structure Of Y281F Mutant Of Hyaluronate Lyase B From Cutibacterium Acnes
Organism: Cutibacterium acnes hl110pa3
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-13 Classification: LYASE |
 |
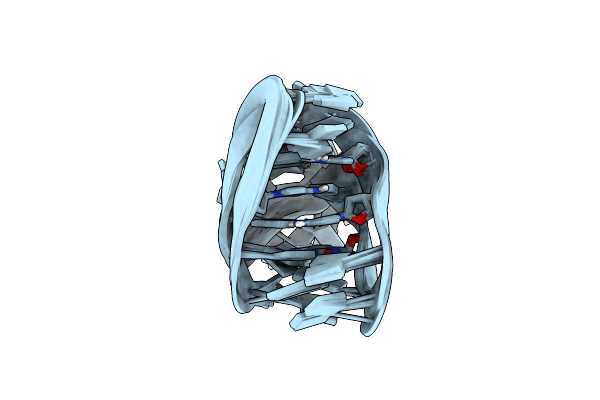 |
An I-Motif Domain Able To Undergo Ph-Dependent Conformational Transitions (Acidic Structure)
|
 |
An I-Motif Domain Able To Undergo Ph-Dependent Conformational Transitions (Neutral Structure)
|

