Search Count: 102
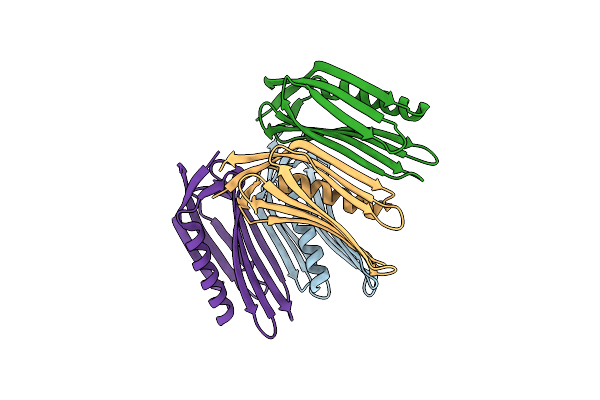 |
Crystal Structure Of A De Novo Designed Monomeric Mainly-Beta Protein B10 (Orthorhombic)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: BIOSYNTHETIC PROTEIN |
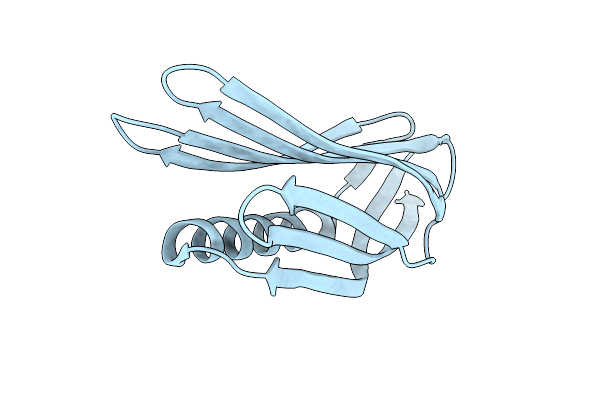 |
Crystal Structure Of A De Novo Designed Monomeric Mainly-Beta Protein B10 (Monoclinic)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: DE NOVO PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Enterobacteria phage t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: EDO, GOL |
 |
Organism: Micromonospora pattaloongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-15 Classification: HYDROLASE |
 |
Organism: Kutzneria buriramensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: ZN, 1PE, ACT |
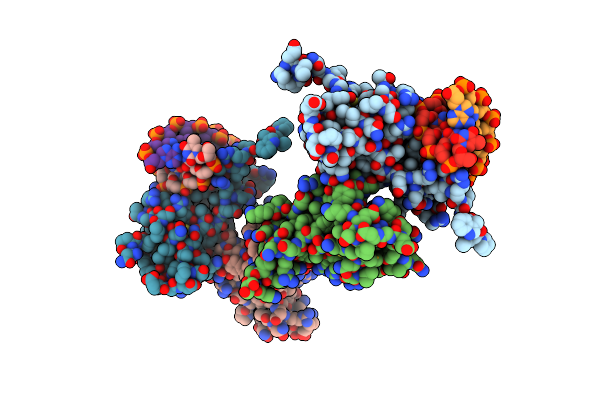 |
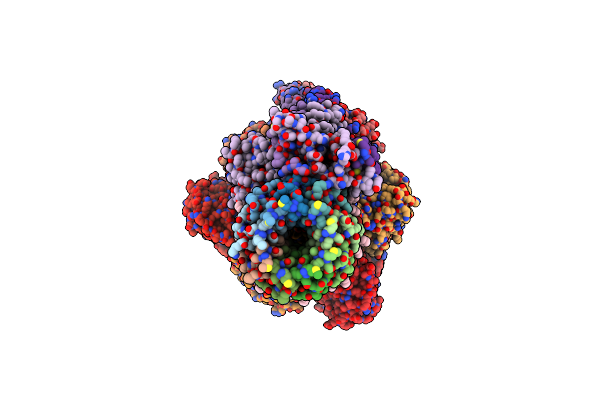
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN |
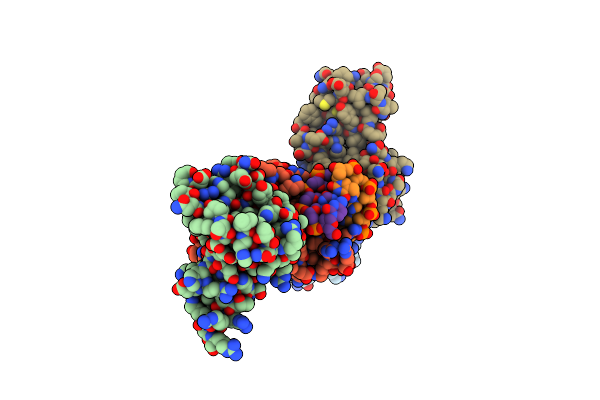 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-09-25 Classification: DNA BINDING PROTEIN |
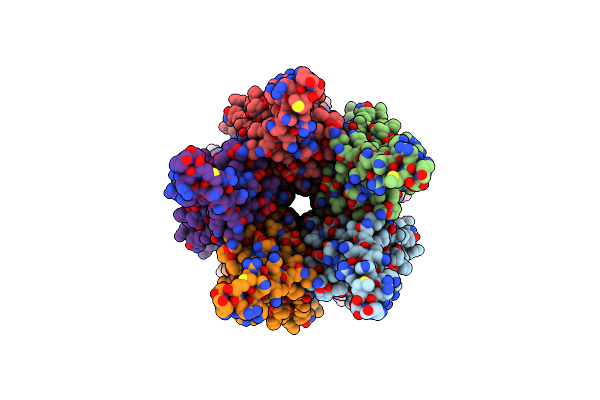 |
Organism: Caenorhabditis elegans, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG, A1D8E, BET |
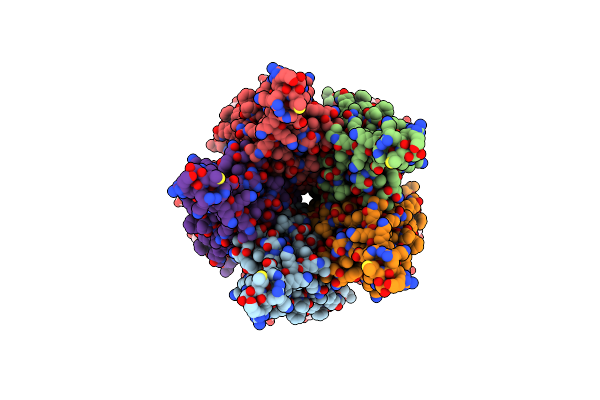 |
Organism: Caenorhabditis elegans, Shimwellia blattae
Method: ELECTRON MICROSCOPY Resolution:2.61 Å Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG |
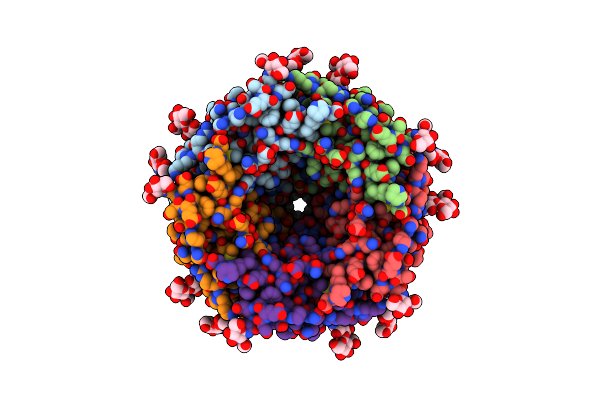 |
Organism: Caenorhabditis elegans, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: TRANSPORT PROTEIN Ligands: NAG, BET |
 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Atp Synthase In Complex With Bedaquiline(Bdq)
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP, BQ1 |
 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Atp Synthase In The Apo-Form
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Atp Synthase Fo In Complex With Bedaquiline(Bdq)
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: BQ1 |
 |
Cryo-Em Structure Of Mycobacterium Tuberculosis Atp Synthase Fo In The Apo-Form
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN |

