Search Count: 31
 |
Organism: Bacillus anthracis, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2019-07-31 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus anthracis, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-07-24 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
 |
X-Ray Structure Of A Non-Autocrystallizing Galectin-10 Variant, Gal10-Tyr69Glu
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: PGE, PG4, P6G |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 6F5
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, PGE |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 1D11
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN Ligands: PG4, ACT |
 |
Structure Of Galectin-10 In Complex With The Fab Fragment Of A Charcot-Leyden Crystal Solubilizing Antibody, 4E8
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2019-06-05 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-06-05 Classification: IMMUNE SYSTEM Ligands: RIP |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: MG, DUP |
 |
Crystal Structure Of Dr2231_E46A Mutant In Complex With Dumpnpp And Manganese
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: MN, GOL, DUP |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: METAL BINDING PROTEIN Ligands: UMP, MG |
 |
Crystal Structure Of Dr2231_E47A Mutant In Complex With Dumpnpp And Magnesium
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: DUP, MG |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: MN, UMP |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: UMP |
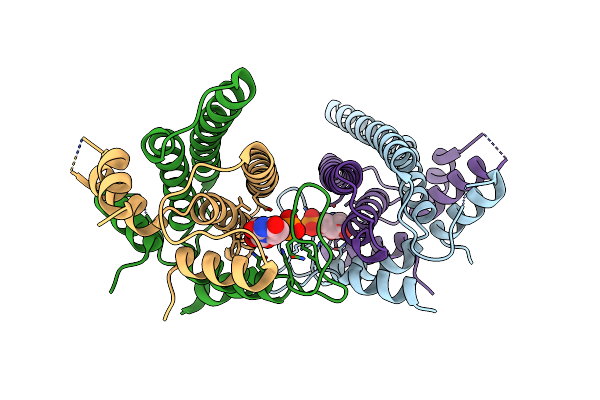 |
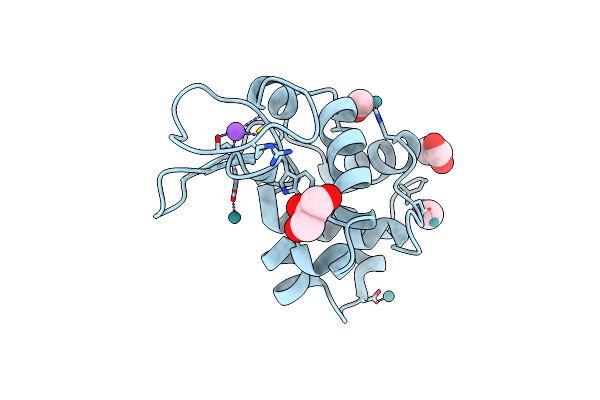
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: UMP |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Methione Oxide Ligand (Complex 6B)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, RU, CMO, GOL |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Pyridine Ligand (Complex 7)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, FMT, CMO, RU |
 |
Lysozyme Soaked With A Ruthenium Based Corm With A Pyridine Ligand (Complex 8)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: CL, NA, RU, CMO |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-10-30 Classification: OXIDOREDUCTASE Ligands: FMN, 12P |

