Search Count: 15
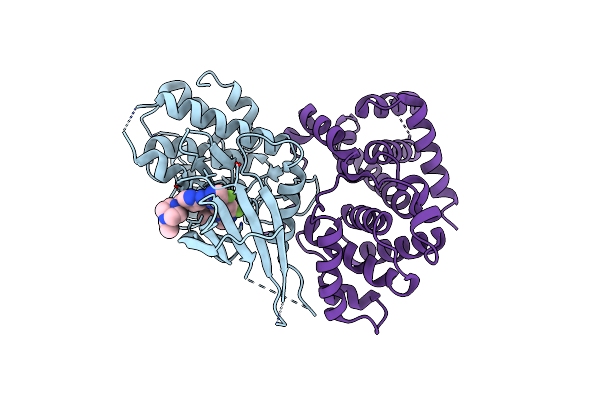 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-11-02 Classification: CELL CYCLE Ligands: 6ZV |
 |
Crystal Structure Of Escherichia Coli Orotate Phosphoribosyltransferase In Complex With Orotic Acid And Sulfate At 1.25 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-11-25 Classification: TRANSFERASE Ligands: ORO, GOL, SO4 |
 |
Crystal Structure Of Escherichia Coli Orotate Phosphoribosyltransferase With An Empty Active Site At 1.55 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: ACT, GOL |
 |
Crystal Structure Of Escherichia Coli Orotate Phosphoribosyltransferase In Complex With Orotic Acid 1.60 Angstrom Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: ORO, GOL |
 |
Organism: Kluyveromyces lactis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-11 Classification: OXIDOREDUCTASE Ligands: HEM, NDP, GOL, K, CL |
 |
Organism: Komagataella pastoris
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-03-04 Classification: OXIDOREDUCTASE Ligands: HEM, NDP, GOL, SO4, CL, K, NA, PEG |
 |
Crystal Structure Of A Streptococcal Dehydrogenase At 1.5 Angstroem Resolution
Organism: Streptococcus pyogenes mgas8232
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, PEG |
 |
Crystal Structure Of A Clostridial Dehydrogenase At 2.55 Angstroems Resolution
Organism: Clostridium perfringens sm101
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, GOL, PEG, ACT |
 |
Structure And Mechanism Of Salicylate Hydroxylase From Pseudomonas Putida G7
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2018-12-26 Classification: OXIDOREDUCTASE Ligands: IOD, GOL, EDO, SO4 |
 |
Crystal Structure Of The Inhibited Form Of The Redox-Sensitive Sufe-Like Sulfur Acceptor Csde From Escherichia Coli At 2.40 Angstrom Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-03-28 Classification: SULFUR-ACCEPTOR PROTEIN Ligands: GOL, SO4 |
 |
Crystal Structure Of A Bacterial Dehydrogenase At 2.19 Angstroms Resolution
Organism: Atopobium vaginae
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: NAD, GOL |
 |
Organism: Fusarium oxysporum
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of A Xylanase In Complex With A Monosaccharide At 2.84 Angstroem Resolution
Organism: Fusarium oxysporum f. sp. vasinfectum 25433
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: GOL, 6MJ |
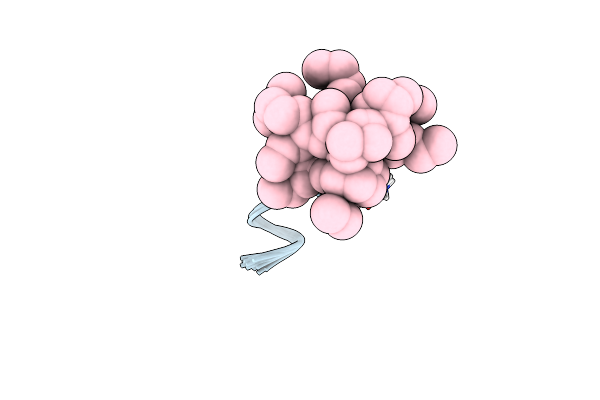 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2007-02-13 Classification: ANTIMICROBIAL PROTEIN Ligands: DAO |
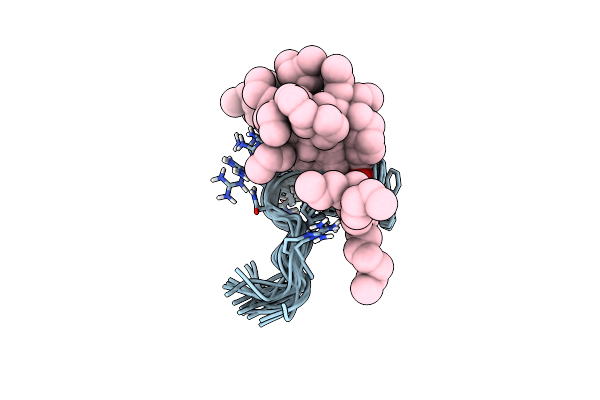 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2007-02-13 Classification: ANTIMICROBIAL PROTEIN Ligands: DAO |

