Search Count: 14
 |
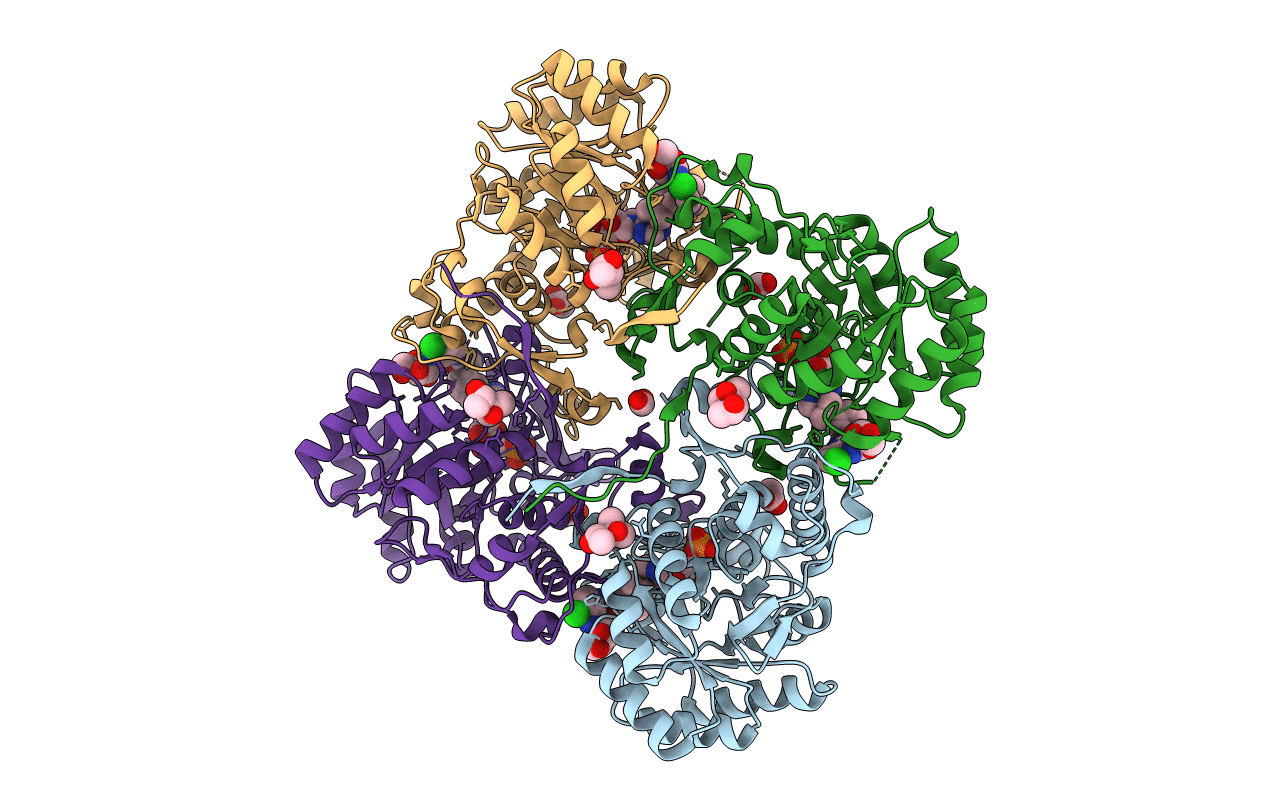
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P225
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2017-05-24 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8KY |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P221
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8N1, MPD, ACY, FMT, MRD |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P182
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L1, K, GOL |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P200
Organism: Clostridium perfringens (strain atcc 13124 / dsm 756 / jcm 1290 / ncimb 6125 / nctc 8237 / type a)
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2017-03-22 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: IMP, 8L4, PGE, PEG, EDO, MG, K, PG4 |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P178
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: IMP, 8LA, K, MPD, FMT, SO4 |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And P176
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-03-01 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: 8L7, IMP, MPD, ACY, MRD |
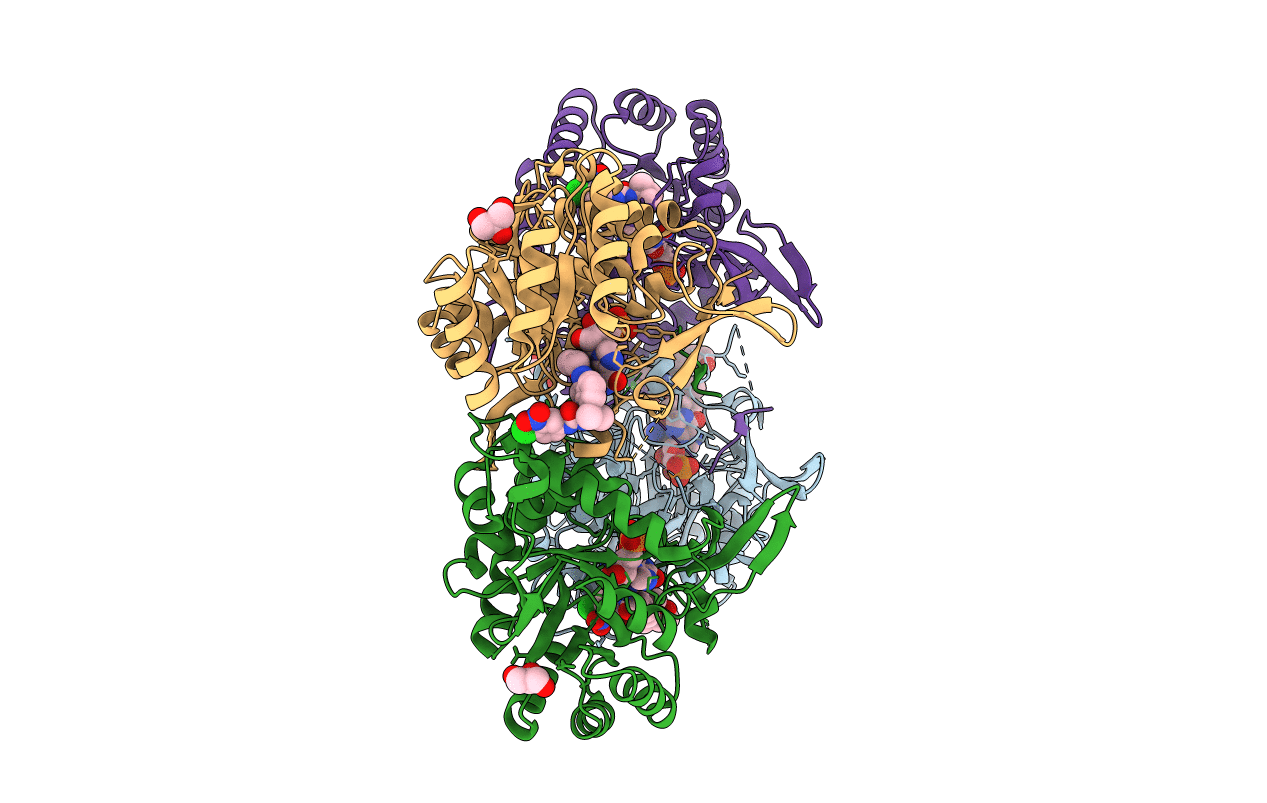 |
Co-Crystal Structure Of The Catalytic Domain Of The Inosine Monophosphate Dehydrogenase From Cryptosporidium Parvum And The Inhibitor P131
Organism: Cryptosporidium parvum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-12-31 Classification: OXIDOREDUCTASE Ligands: IMP, I13, GOL, PEG |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And C91
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2014-07-09 Classification: OXIDOREDUCTASE Ligands: IMP, C91 |
 |
Crystal Structure Of Inosine 5'-Monophosphate Dehydrogenase From Clostridium Perfringens Complexed With Imp And A110
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2014-07-09 Classification: OXIDOREDUCTASE Ligands: IMP, 2YA, ACY, FMT, SO4, GOL |
 |
Crystal Structure Of Bacillus Anthracis Inosine Monophosphate Dehydrogenase In The Complex With Imp
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2011-12-07 Classification: OXIDOREDUCTASE Ligands: IMP, SO4, GOL, CL |
 |
Crystal Structure Of Inosine-5'-Monophosphate Dehydrogenase From Bacillus Anthracis Str. Ames
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-10-05 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Inosine-5'-Monophosphate Dehydrogenase From Bacillus Anthracis Str. Ames Complexed With Xmp
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-10-05 Classification: OXIDOREDUCTASE Ligands: XMP, TAR, SO4 |
 |
Crystal Structure Of Human Guanosine Monophosphate Reductase 2 Gmpr2 In Complex With Imp And Nadph
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-11-30 Classification: OXIDOREDUCTASE Ligands: IMP, NDP |
 |
Crystal Structure Of Human Guanosine Monophosphate Reductase 2 Gmpr2 In Complex With Imp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2005-09-02 Classification: OXIDOREDUCTASE Ligands: IMP |

