Search Count: 109
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: PROTEIN TRANSPORT Ligands: MAN, NAG |
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Zoledronate
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: ZOL, MG, LMT |
 |
Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima In Complex With Ca2+ And Etidronate
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: MEMBRANE PROTEIN Ligands: 911, MG, CA, LMT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-05-22 Classification: IMMUNE SYSTEM Ligands: FLC, PO4, 16P, PGE, DXE, FWN, NA |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 0-60-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: LMT, PEG, PGE, PG4 |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 300-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.98 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG |
 |
Time-Resolved Structure Of K+-Dependent Na+-Ppase From Thermotoga Maritima 3600-Seconds Post Reaction Initiation With Na+
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:4.53 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: DPO, MG, K, PO4 |
 |
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-12-20 Classification: TRANSFERASE Ligands: UDP, VVC |
 |
C-Terminal Head Domain Of The Trimeric Autotransporter Adhesin Bpac From Burkholderia Pseudomallei Fused To A Gcn4 Anchor
Organism: Burkholderia pseudomallei 1026b, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-06-29 Classification: CELL ADHESION Ligands: GOL, NA |
 |
Crystal Structure Of Complex Between Fh19-20 And Fhba Protein From Borrelia Hermsii
Organism: Borrelia hermsii yor, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-01-12 Classification: IMMUNE SYSTEM Ligands: EDO, SO4 |
 |
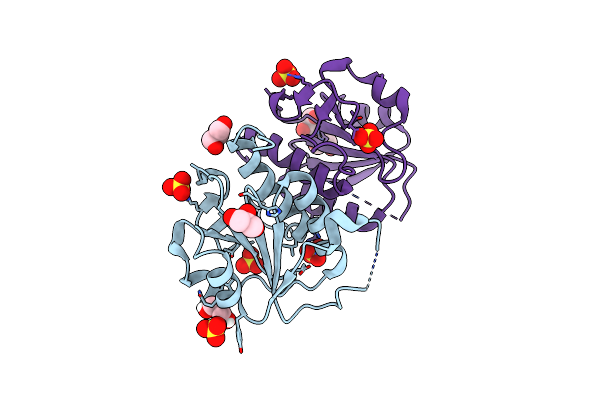
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: SO4, PEG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-02-19 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
Structure Of 299-452 Fragment Of The Uspa1 Protein From Moraxella Catarrhalis
Organism: Moraxella catarrhalis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-08-21 Classification: CELL ADHESION Ligands: CL, HEZ, ZN |
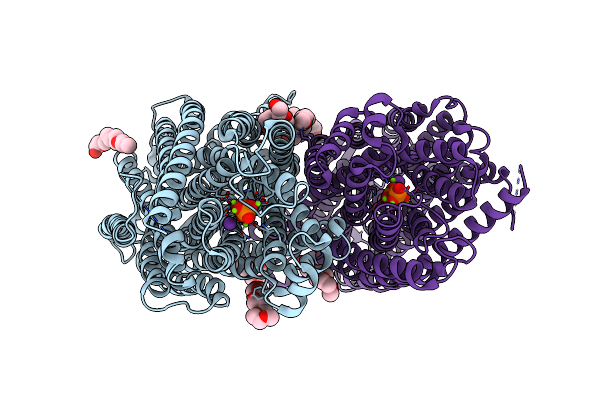 |
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: PO4, MG, K, 1PG |
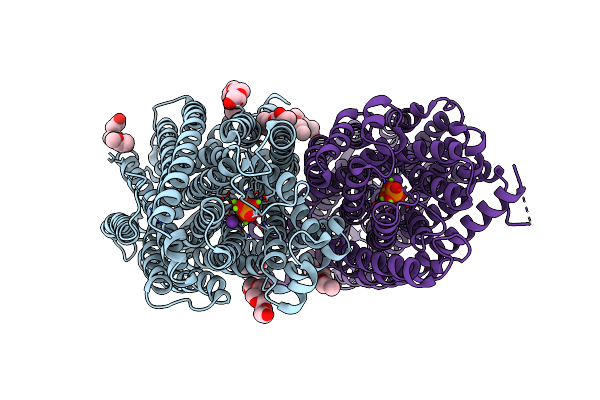 |
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: PO4, MG, K, 1PG |
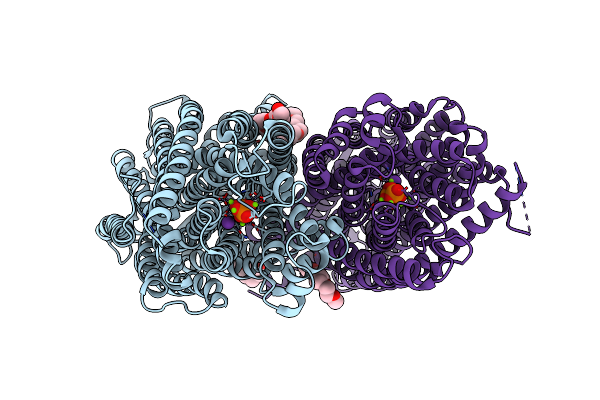 |
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: PO4, MG, K, 1PG |
 |
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: PO4, MG, K, 1PG |
 |
Organism: Vigna radiata var. radiata
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: PO4, MG, K, 1PG |

