Search Count: 20
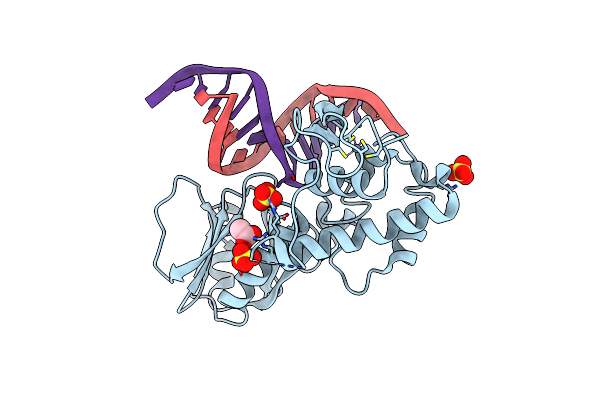 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (E2Q) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: SO4, ZN, ACT |
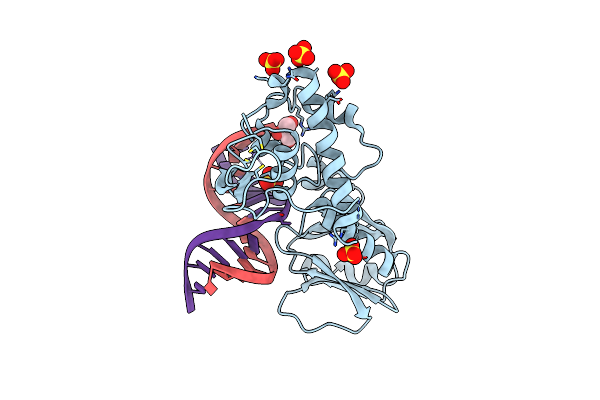 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA Ligands: SO4, ZN, GOL |
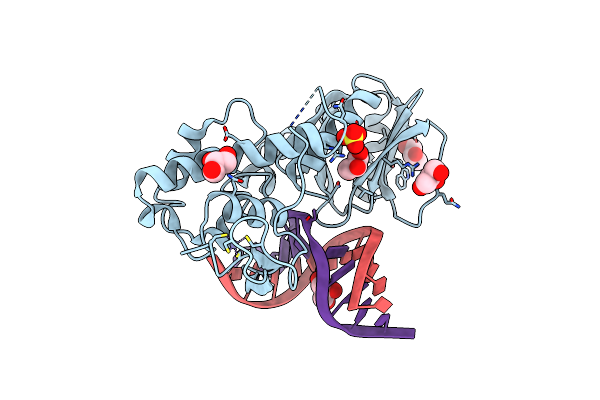 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (R252A) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA Ligands: GOL, ZN, SO4 |
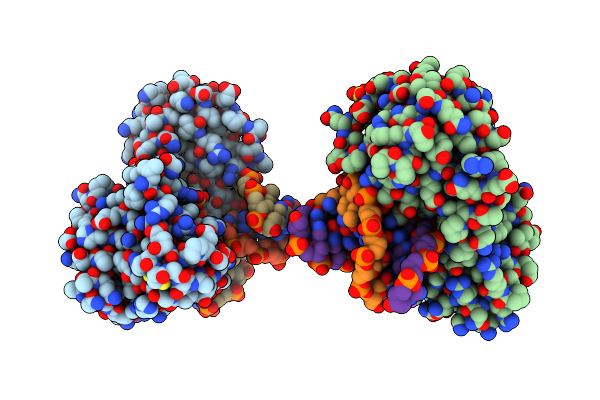 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (E2Q) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA |
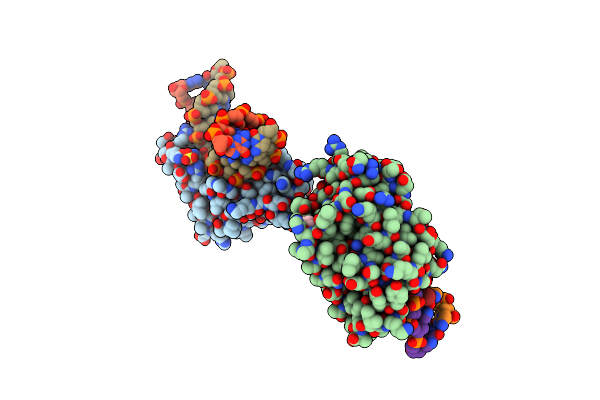 |
Crystal Structure Of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed With A Dna Substrate Containing Abasic Site
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-10-03 Classification: HYDROLASE Ligands: SO4, GOL |
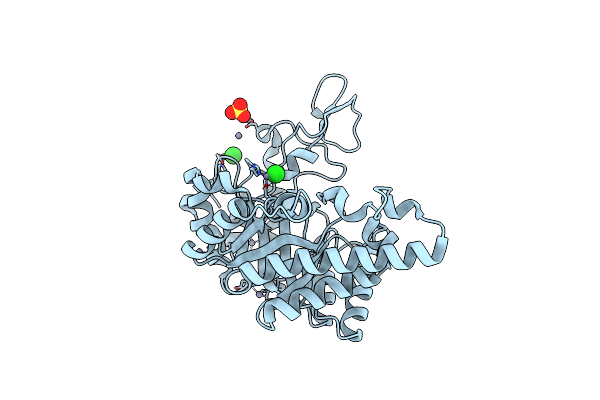 |
Crystal Structure Of The Extracellular Xylanase From Geobacillus Stearothermophilus T-6 (Xt6, Monoclinic Form): The E159A/E265A Mutant At 1.8A Resolution
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-07-19 Classification: HYDROLASE Ligands: ZN, CL, SO4 |
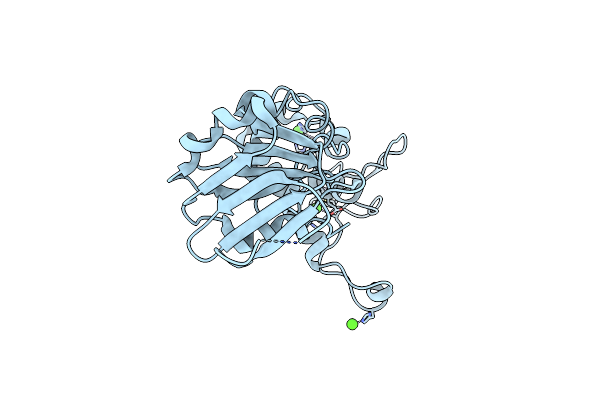 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The Wt Enzyme At 2.8 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, CA |
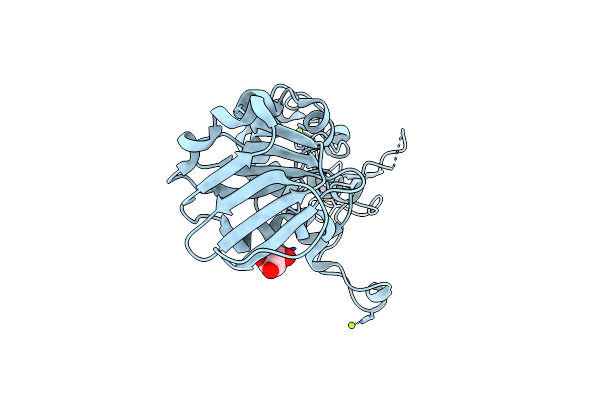 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The R252A Mutant At 2.05 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The E2A Mutant At 2.3 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
 |
The High-Resolution Crystal Structure Of Ixt6, A Thermophilic, Intracellular Xylanase From G. Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2003-11-25 Classification: HYDROLASE |
 |
The Crystal Structure Of Alpha-D-Glucuronidase From Bacillus Stearothermophilus T-1 Complexed With Glucuronic Acid
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-09-17 Classification: HYDROLASE Ligands: GCU, GOL |
 |
The Crystal Structure Of Alpha-D-Glucuronidase (E386Q) From Bacillus Stearothermophilus T-6
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-09-17 Classification: HYDROLASE Ligands: GOL |
 |
The 1.5A Crystal Structure Of Alpha-D-Glucuronidase From Bacillus Stearothermophilus T-1, Complexed With 4-O-Methyl-Glucuronic Acid And Xylotriose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2003-03-21 Classification: HYDROLASE Ligands: GCW, GOL |
 |
The 1.7 A Crystal Structure Of Alpha-D-Glucuronidase, A Family-67 Glycoside Hydrolase From Bacillus Stearothermophilus T-1
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2002-10-29 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of A Mutated Family-67 Alpha-D-Glucuronidase (E285N) From Bacillus Stearothermophilus T-6, Complexed With 4-O-Methyl-Glucuronic Acid
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2002-10-29 Classification: HYDROLASE Ligands: GCV, GOL |
 |
Crystal Structure Of A Mutated Family-67 Alpha-D-Glucuronidase (E285N) From Bacillus Stearothermophilus T-6, Complexed With Aldotetraouronic Acid
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2002-10-29 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4 |
 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Brominated-Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4, GOL |
 |
Crystal Structure Of E.Coli Formamidopyrimidine-Dna Glycosylase (Fpg) Covalently Trapped With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-06-14 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-01-04 Classification: GLYCOSIDASE Ligands: GLA, GLC, SO4 |

