Search Count: 8
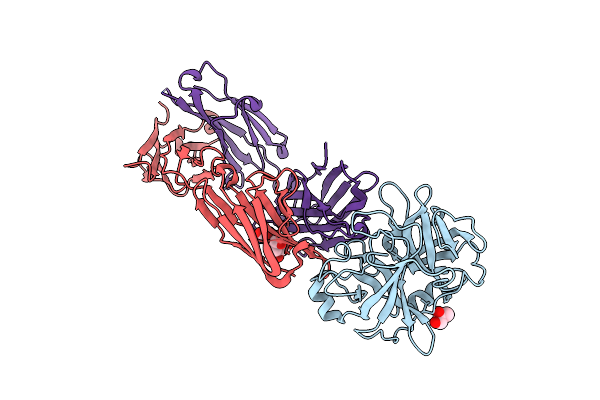 |
Structures Of Fab-Protease Complexes Reveal A Highly Specific Non-Canonical Mechanism Of Inhibition.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-06-20 Classification: HYDROLASE Ligands: GOL |
 |
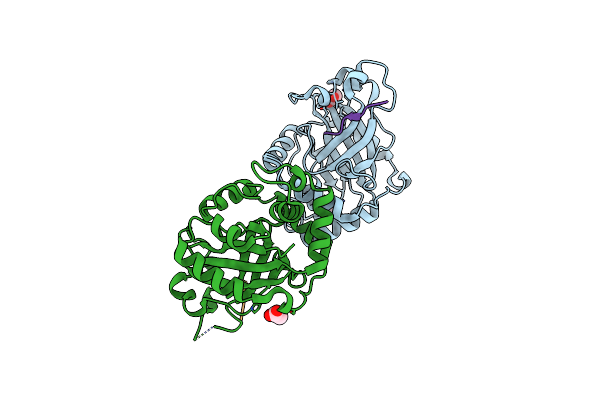
Crystal Structure Of Membrane-Type Serine Protease 1 (Mt-Sp1) In Complex With The Fab Inhibitor S4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2011-07-06 Classification: HYDROLASE/IMMUNE SYSTEM Ligands: EDO, NA, CL |
 |
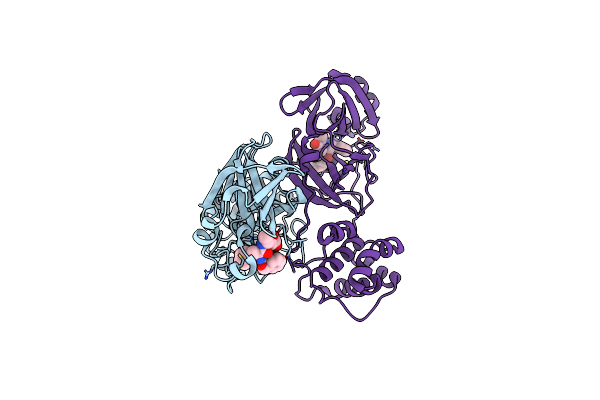
Crystal Structure Of Kshv Protease In Complex With Hexapeptide Phosphonate Inhibitor
Organism: Human herpesvirus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2007-12-25 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ACT |
 |
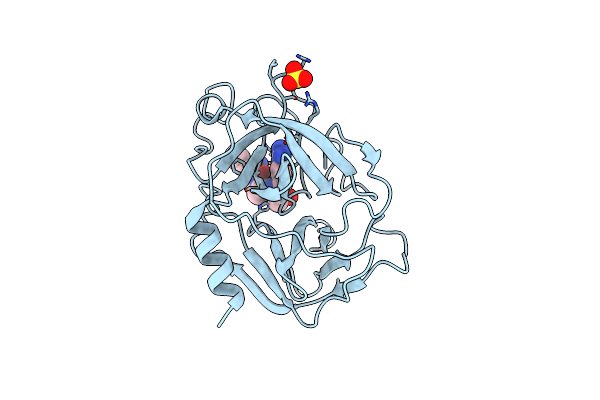
Substrate Specificity Profiling And Identification Of A New Class Of Inhibitor For The Major Protease Of The Sars Coronavirus
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: WR1 |
 |
The Oligomeric Structure Of Human Granzyme A Reveals The Molecular Determinants Of Substrate Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-07-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0G6, SO4 |
 |
Neutrophil Gelatinase-Associated Lipocalin Is A Novel Bacteriostatic Agent That Interferes With Siderophore-Mediated Iron Acquisition
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2003-03-11 Classification: TRANSPORT PROTEIN Ligands: FE, SO4, DBH, DBS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2000-04-21 Classification: SUGAR BINDING PROTEIN Ligands: DKA |
 |
Crystal Structure Of Human Neutrophil Gelatinase Associated Lipocalin Monomer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2000-03-06 Classification: SUGAR BINDING PROTEIN Ligands: SO4, NAG |

