Search Count: 198
 |
High Resolution Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: FMT, SO4 |
 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Avibactam
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: NXL, SIN, FMT, EDO |
 |
Structure Of Class A Beta-Lactamase From Bordetella Bronchiseptica Rb50 In A Complex With Clavulonate
Organism: Bordetella bronchiseptica rb50
Method: X-RAY DIFFRACTION Release Date: 2024-06-12 Classification: HYDROLASE Ligands: MLA, TEM, CL |
 |
Crystal Structure Of The Second Form Of The Co-Factor Complex Of Nsp7 And The C-Terminal Domain Of Nsp8 From Sars Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-13 Classification: VIRAL PROTEIN Ligands: ACY |
 |
Crystal Structure Of Motility Associated Killing Factor B From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2020-03-25 Classification: TOXIN Ligands: EDO |
 |
Crystal Structure Of The Beta Lactamase Class A Penp From Bacillus Subtilis In The Complex With The Non-Beta- Lactam Beta-Lactamase Inhibitor Avibactam
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Release Date: 2020-03-25 Classification: HYDROLASE Ligands: NXL, FMT, EDO |
 |
Crystal Structure Of The Methyltransferase-Stimulatory Factor Complex Of Nsp16 And Nsp10 From Sars Cov-2.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-25 Classification: VIRAL PROTEIN Ligands: CL, EDO, SAM, ZN |
 |
The 1.9 A Crystal Structure Of Nsp15 Endoribonuclease From Sars Cov-2 In The Complex With A Citrate
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-03-11 Classification: VIRAL PROTEIN Ligands: EDO, PEG, CIT |
 |
Crystal Structure Of Motility Associated Killing Factor E From Vibrio Cholerae
Organism: Vibrio cholerae o1 biovar el tor str. n16961
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TOXIN Ligands: ACY, EDO, K, CL, FMT |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-03-04 Classification: VIRAL PROTEIN Ligands: GOL, MG, ACY, CL |
 |
2.75 Angstrom Crystal Structure Of Galactarate Dehydratase From Escherichia Coli.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-11-06 Classification: LYASE Ligands: CA, CL |
 |
The 2.0 A Crystal Structure Of The Type B Chloramphenicol Acetyltransferase From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: MPD, EDO, CL, PO4 |
 |
The Crystal Structure Of Chloramphenicol Acetyltransferase-Like Protein From Vibrio Fischeri Es114 In Complex With Taurocholic Acid
Organism: Aliivibrio fischeri (strain atcc 700601 / es114)
Method: X-RAY DIFFRACTION Release Date: 2019-09-25 Classification: TRANSFERASE Ligands: TCH, CL, SO4, ACT, GOL, FMT |
 |
Crystal Structure Of The Type B Chloramphenicol O-Acetyltransferase From Vibrio Vulnificus
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Release Date: 2019-08-14 Classification: TRANSFERASE Ligands: EDO, CL |
 |
2.6 Angstrom Resolution Crystal Structure Of Aminopeptidase B From Escherichia Coli Str. K-12 Substr. Mg1655
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-05-15 Classification: HYDROLASE Ligands: ZN, MN, CL |
 |
2.05 Angstrom Resolution Crystal Structure Of Aminopeptidase B From Escherichia Coli Str. K-12 Substr. Mg1655.
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: ZN, CA, CL, BCT, EDO, PGE, PEG |
 |
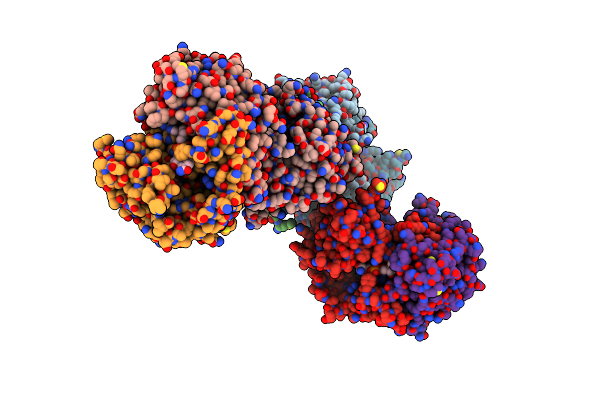
Crystal Structure Of A Tripartite Toxin Component Vca0883 From Vibrio Cholerae
Organism: Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE |
 |
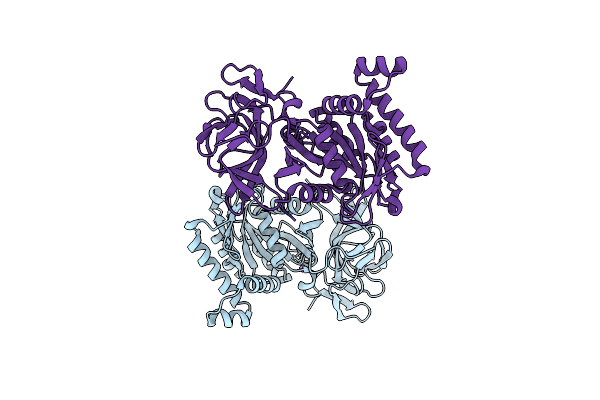
Crystal Structure Of Nad Synthetase (Nade) From Enterococcus Faecalis In Complex With Nad+
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Release Date: 2018-02-14 Classification: TRANSFERASE Ligands: NAD |
 |
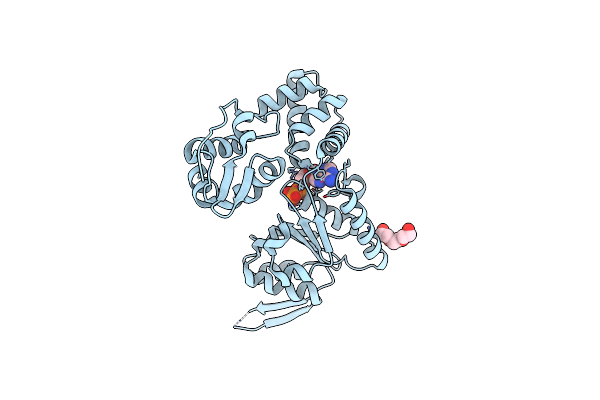
1.83 Angstrom Resolution Crystal Structure Of N-Terminal Fragment (Residues 1-404) Of Elongation Factor G From Enterococcus Faecalis
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Release Date: 2017-11-22 Classification: RIBOSOMAL PROTEIN |
 |
1.88 Angstrom Resolution Crystal Structure Holliday Junction Atp-Dependent Dna Helicase (Ruvb) From Pseudomonas Aeruginosa In Complex With Adp
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Release Date: 2017-11-22 Classification: HYDROLASE Ligands: ADP, PGE |

