Search Count: 135
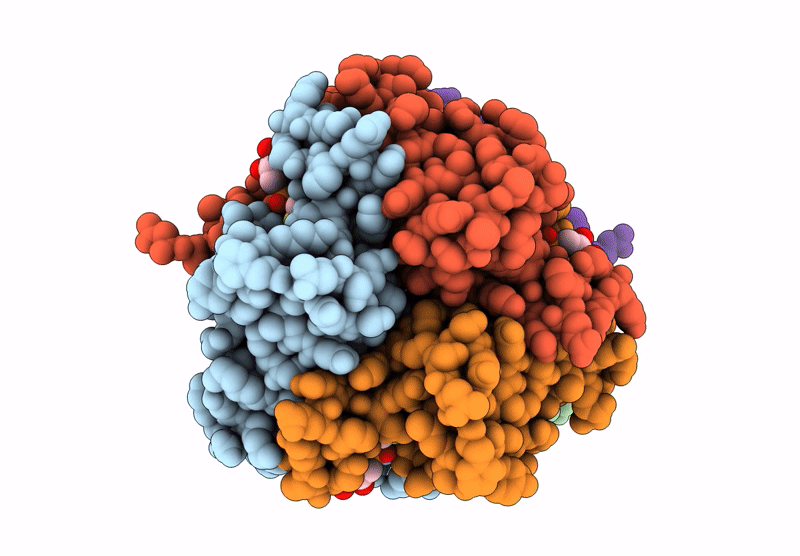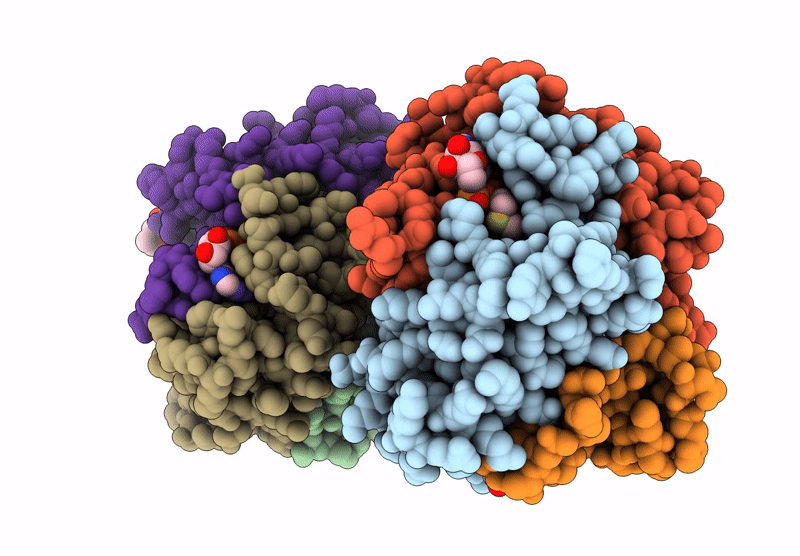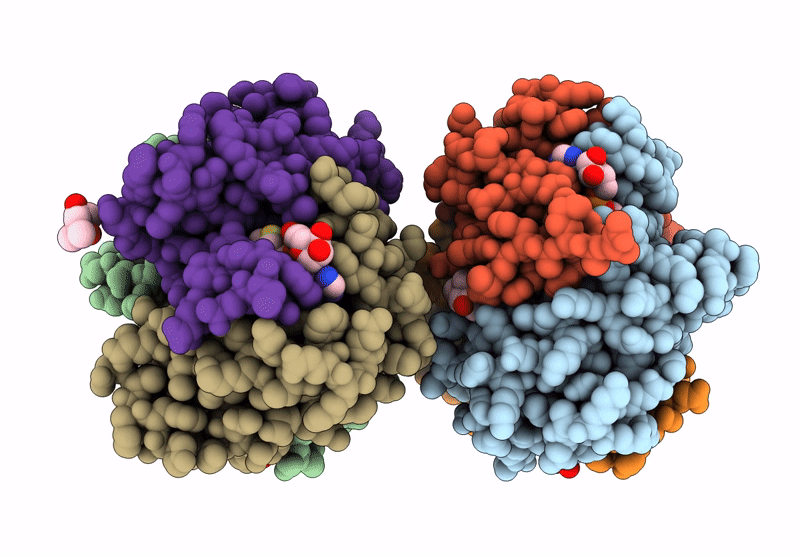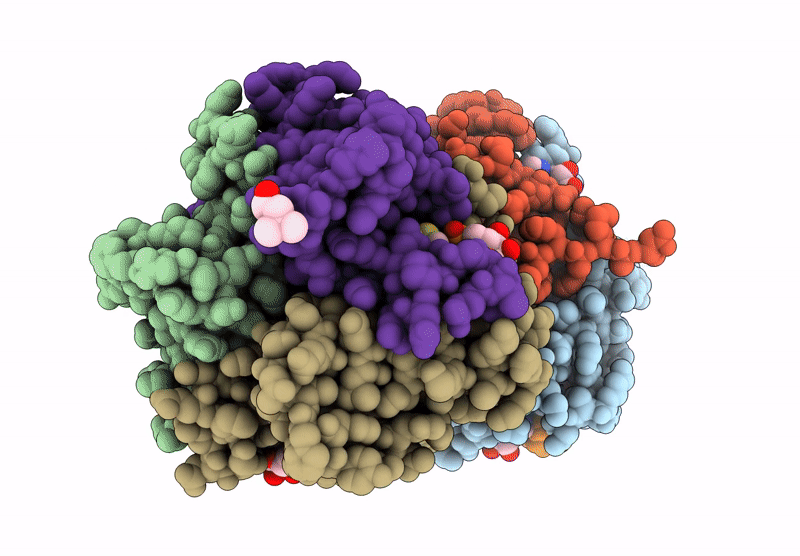 |
Structure And Catalytic Mechanism Of Sam-Amp Lyase In Clostridium Botulinum Cora-Associated Type Iii Crispr System
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: A1IJB, MPD |
 |
Structure And Catalytic Mechanism Of Sam-Amp Lyase In Clostridium Botulinum Cora-Associated Type Iii Crispr System
Organism: Clostridium botulinum
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: LYASE Ligands: MPD |
 |
A Viral Saved Protein With Ring Nuclease Activity Subverts Type Iii Crispr Defence
Organism: Thermocrinis great boiling spring virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: VIRAL PROTEIN Ligands: PO4 |
 |
Structure Of The Cytoplasmic Domain Of Csx23 From Vibrio Cholera In Complex With Cyclic Tetra-Adenylate (Ca4)
Organism: Vibrio cholerae, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-08-07 Classification: GENE REGULATION Ligands: ACE, NA |
 |
Crystal Structure Of Beta-Xylosidase Mutant (D281N, E517Q) From Thermotoga Maritima In Complex With Xylopentaose
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-01-24 Classification: HYDROLASE Ligands: MPD |
 |
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
Structure Of Csm6' From Streptococcus Thermophilus In Complex With Cyclic Hexa-Adenylate (Ca6)
Organism: Streptococcus thermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
Crystal Structure Of Beta-Xylosidase From Thermotoga Maritima In Complex With Methyl-Beta-D-Xylopyranoside
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: MPD, 6MJ |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: MPD |
 |
Crystal Structure Of Beta-Xylosidase From Thermotoga Maritima In Complex With Methyl-Beta-D-Xylopyranoside Hydrolysed To Xylose
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: XYP, MPD |
 |
Crystal Structure Of Beta-Xylosidase From Thermotoga Maritima In Complex With Xylohexaose Hydrolysed To Xylobiose
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-04-05 Classification: HYDROLASE Ligands: MPD, IVG |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-04-27 Classification: RNA BINDING PROTEIN Ligands: GOL, CIT |
 |
Structure Of Staphylococcus Aureus M1A22-Trna Methyltransferase In Complex With S-Adenosylmethionine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-04-20 Classification: RNA BINDING PROTEIN Ligands: SAM, GOL |
 |
Structure Of Staphylococcus Aureus M1A22-Trna Methyltransferase In Complex With S-Adenosylhomocysteine
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-04-06 Classification: RNA BINDING PROTEIN Ligands: SAH, GOL |
 |
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KDM, CA |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2,3-Difluoro-2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: FKD, K99, GOL, CL, CA |
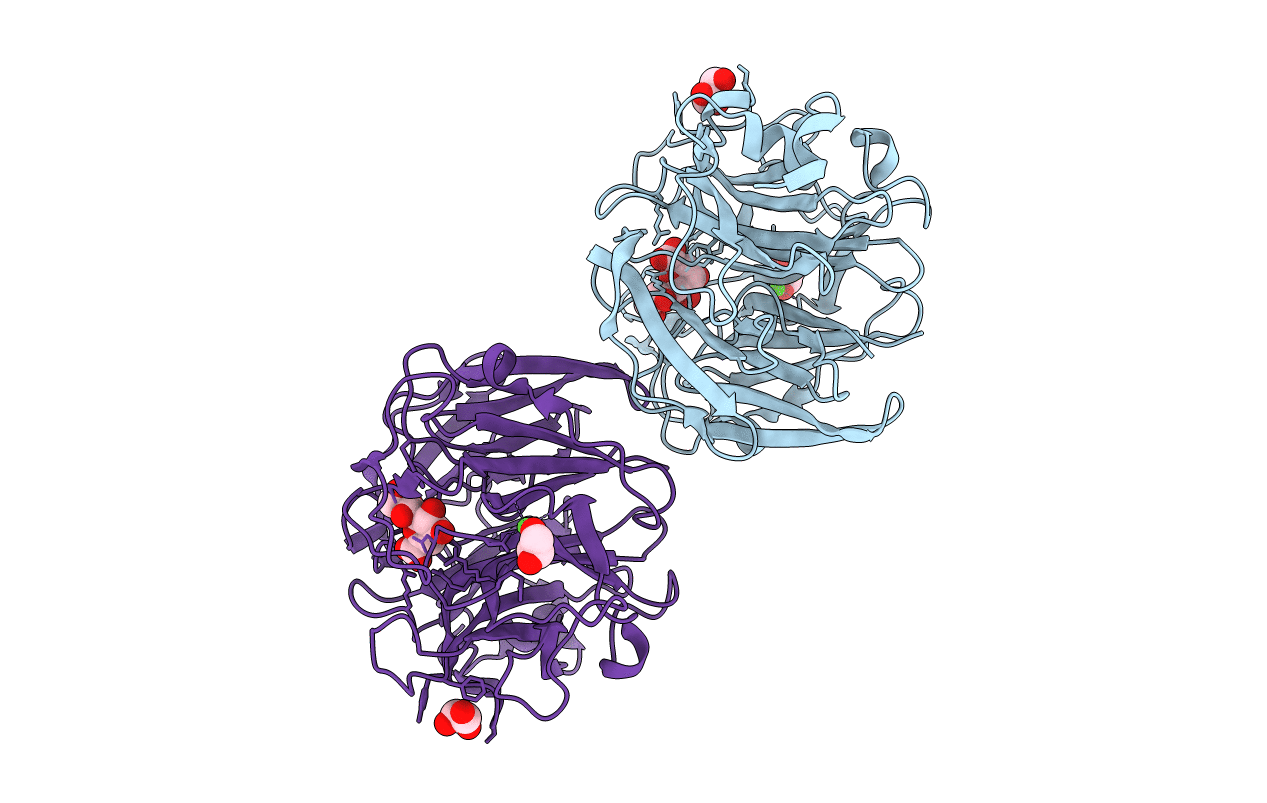 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2,3-Didehydro-2,3-Dideoxy-D-Glycero-D-Galacto-Nonulosonic Acid.
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KFN, GOL, CA |
 |
Structure Of Kdnase From Aspergillus Terrerus In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: GOL, KDM, CL |
 |
Structure Of Kdnase From Trichophyton Rubrum In Complex With 2-Keto-3-Deoxynononic Acid
Organism: Trichophyton rubrum
Method: X-RAY DIFFRACTION Resolution:0.91 Å Release Date: 2021-10-20 Classification: CARBOHYDRATE Ligands: KDM, GOL |

