Search Count: 17
 |
Organism: Hypoxylon sp. ec38
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-09-15 Classification: OXIDOREDUCTASE Ligands: MG, NAG, HEM, IMD, BU1, PEG |
 |
Unspecific Peroxygenase From Hypoxylon Sp. Ec38 In Complex With 1-Phenylimidazole
Organism: Hypoxylon sp. ec38
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-09-15 Classification: OXIDOREDUCTASE Ligands: MG, NAG, HEM, PEG, PIW, GOL |
 |
Unspecific Peroxygenase From Hypoxylon Sp. Ec38 In Complex With S-1,2-Propanediol
Organism: Hypoxylon sp. ec38
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-15 Classification: OXIDOREDUCTASE Ligands: HEM, PGO, NAG, PEG, 1BO, BCN, MG |
 |
Unspecific Peroxygenase From Hypoxylon Sp. Ec38 In Complex With 2-(N-Morpholino) Ethanesulfonic Acid (Mes)
Organism: Hypoxylon sp. ec38
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2021-09-15 Classification: OXIDOREDUCTASE Ligands: MES, NAG, HEM, MG |
 |
Organism: Hypoxylon sp. ec38
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-15 Classification: OXIDOREDUCTASE Ligands: HEM, NAG, DMS, SYN, MG |
 |
Organism: Pseudomonas sp. 19-rlim
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: HEM, SRT, PGE, GOL |
 |
Organism: Phenylobacterium zucineum (strain hlk1)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: HEM |
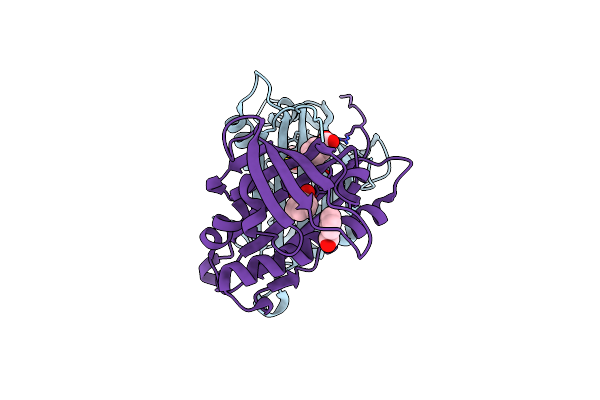 |
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With Benzoic Acid
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-10-12 Classification: LYASE Ligands: BEZ |
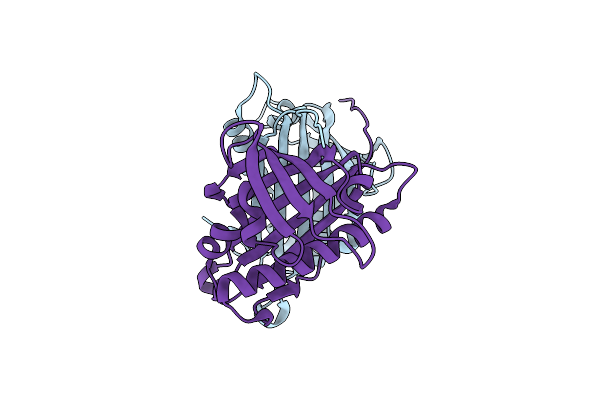 |
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-10-05 Classification: LYASE |
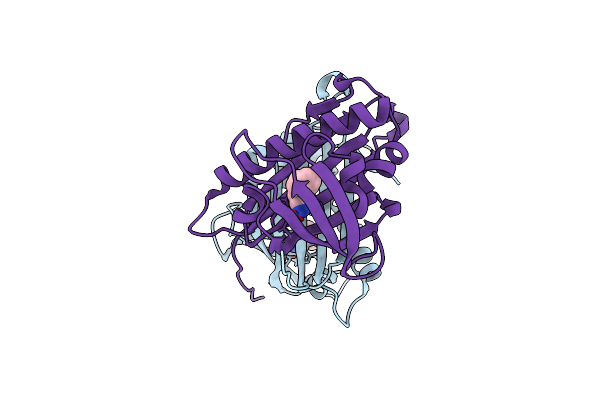 |
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With (R)-Mandelonitrile / Benzaldehyde
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-10-05 Classification: LYASE Ligands: HBX, MXN |
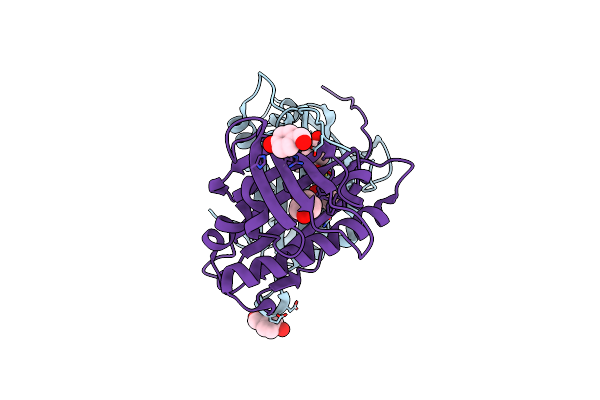 |
Hydroxynitrile Lyase From The Fern Davallia Tyermanii In Complex With P-Hydroxybenzaldehyde
Organism: Davallia tyermannii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-05 Classification: LYASE Ligands: HBA |
 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-03-31 Classification: OXIDOREDUCTASE Ligands: FMN, SO4 |
 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-03-31 Classification: OXIDOREDUCTASE Ligands: FMN, SO4, BTB |
 |
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2009-03-24 Classification: LYASE Ligands: FAD, NAG, HBX, MXN |
 |
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2009-03-24 Classification: LYASE Ligands: IPA, FAD, NAG, NDG |
 |
Organism: Prunus dulcis
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2002-09-04 Classification: LYASE Ligands: NAG, FAD, IPA |

