Search Count: 22
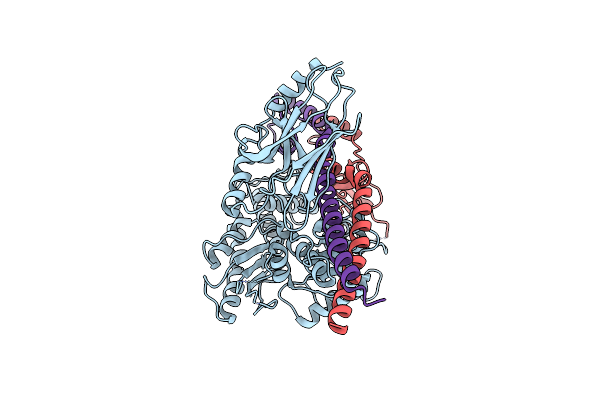 |
Structure Of The Mycobacterium Tuberculosis Hsp70 Protein Dnak Bound To The Nucleotide Exchange Factor Grpe
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: CHAPERONE |
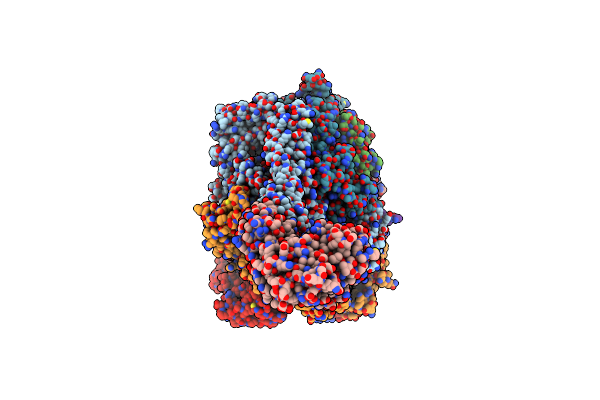 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With A Locally Refined Clpb Middle Domain And A Dnak Nucleotide Binding Domain
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
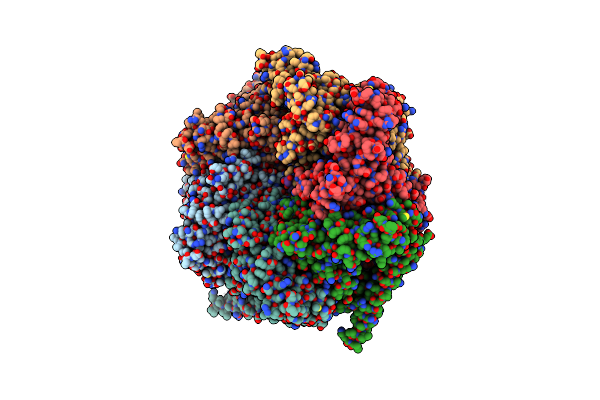 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure In Conformation Ii In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
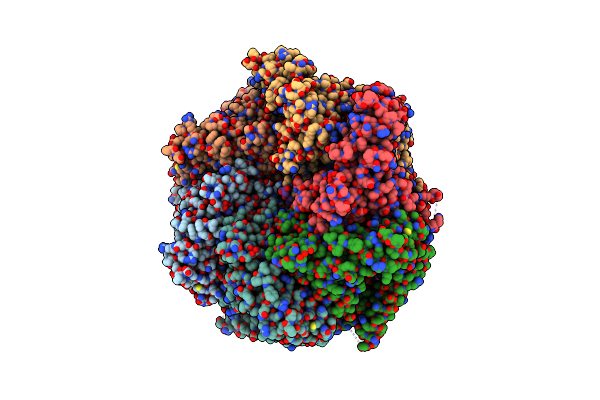 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure In Conformation T In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With A Locally Refined N-Terminal Domain In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure With Three Locally Refined Clpb Middle Domains And Three Dnak Nucleotide Binding Domains
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-05-26 Classification: CHAPERONE Ligands: AGS, ADP |
 |
The Mycobacterium Tuberculosis Clpb Disaggregase Hexamer Structure In Conformation I In The Presence Of Dnak Chaperone And A Model Substrate
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: CHAPERONE Ligands: AGS, ADP |
 |
Crystal Structure Of A Mycobacterium Smegmatis Transcription Initiation Complex With Rifampicin-Resistant Rna Polymerase And Bound To Kanglemycin A
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2018-09-05 Classification: transcription/dna/antibiotic Ligands: KNG, SO4, ZN, MG, GLU, EDO |
 |
Crystal Structure Of A Mycobacterium Smegmatis Rna Polymerase Transcription Initiation Complex With Inhibitor Kanglemycin A
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis str. mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-08-15 Classification: transcription/dna/antibiotic Ligands: KNG, SO4, ZN, MG, GLU, EDO |
 |
Crystal Structure Of A Mycobacterium Smegmatis Rna Polymerase Transcription Initiation Complex With Inhibitor Rifampicin
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Mycobacterium smegmatis str. mc2 155, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2018-08-15 Classification: TRANSCRIPTION Ligands: SO4, EDO, RFP, ZN, GLU |
 |
Crystal Structure Of A Mycobacterium Smegmatis Transcription Initiation Complex With Rbpa
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155), Aquifex aeolicus, Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2017-01-18 Classification: TRANSCRIPTION ACTIVATOR/TRANSFERASE/DNA Ligands: SO4, EDO, ZN, MG |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2013-07-31 Classification: TRANSCRIPTION |
 |
Structure Of Mycobacterium Tuberculosis Mycolic Acid Cyclopropane Synthase Cmaa2 In Complex With Dioctylamine
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2009-06-16 Classification: TRANSFERASE Ligands: D22, CO3 |
 |
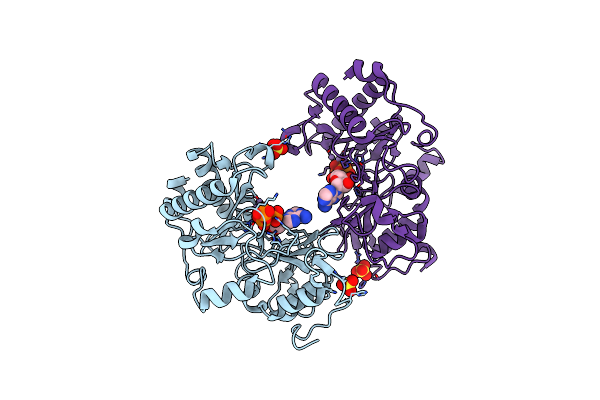
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2006-05-23 Classification: HYDROLASE/TRANSFERASE Ligands: SO4 |
 |
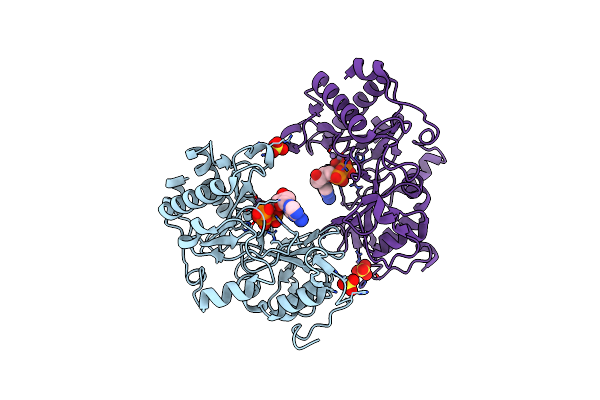
Crystal Structure Of Pseudomonas Aeruginosa Ligd Polymerase Domain With Atp And Manganese
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-05-23 Classification: HYDROLASE/TRANSFERASE Ligands: MN, SO4, ATP |
 |
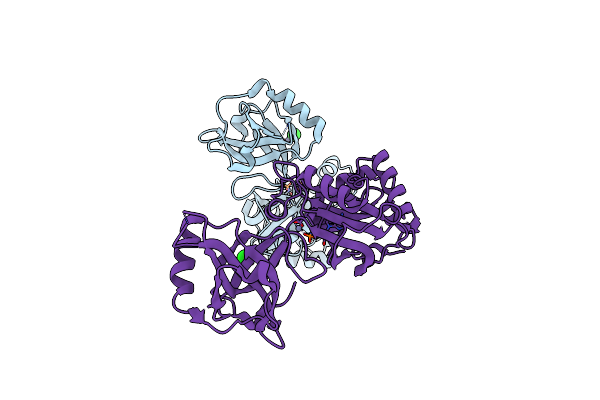
Crystal Structure Of Pseudomonas Aeruginosa Ligd Polymerase Domain With Datp And Manganese
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-05-23 Classification: Hydrolase/Transferase Ligands: MN, SO4, DTP |
 |
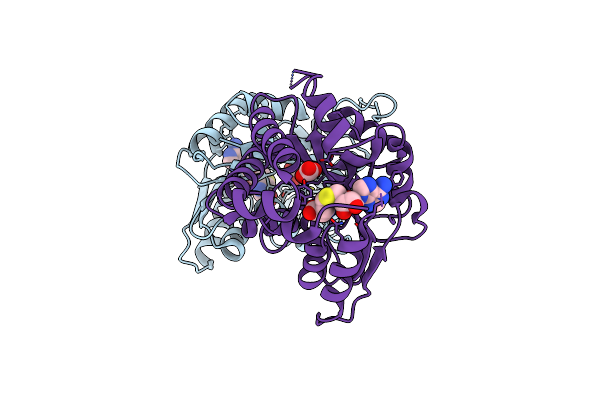
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-02-28 Classification: LIGASE Ligands: ZN, CL, MG |
 |
Crystal Structure Of Mycolic Acid Cyclopropane Synthase Pcaa Complexed With S-Adenosyl-L-Homocysteine
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-03-06 Classification: TRANSFERASE Ligands: CO3, SAH |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2002-01-11 Classification: TRANSFERASE Ligands: ACY |
 |
Crystal Structure Of Mycolic Acid Cyclopropane Synthase Cmaa1 Complexed With Sah And Ctab
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-01-11 Classification: TRANSFERASE Ligands: CO3, SAH, 16A |

