Search Count: 8
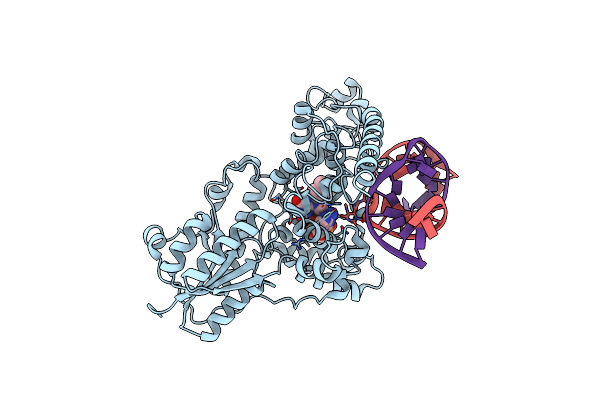 |
Structure Of The 6-4 Photolyase Of D. Melanogaster In Complex With The Non-Natural N4-Methyl T(Dewar)C Lesion
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-02-23 Classification: LYASE/DNA Ligands: FAD |
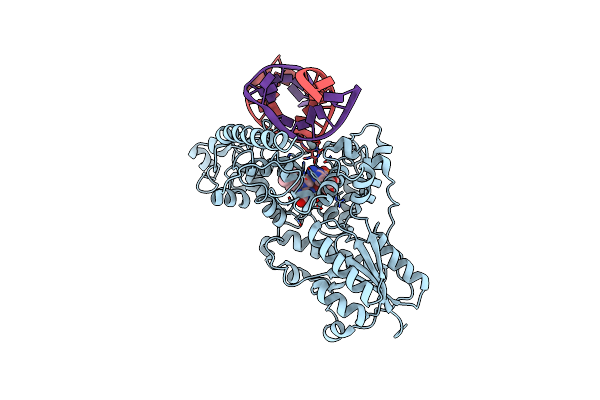 |
Structure Of The 6-4 Photolyase Of D. Melanogaster In Complex With The Non-Natural N4-Methyl T(6-4)C Lesion
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-02-23 Classification: LYASE/DNA Ligands: FAD |
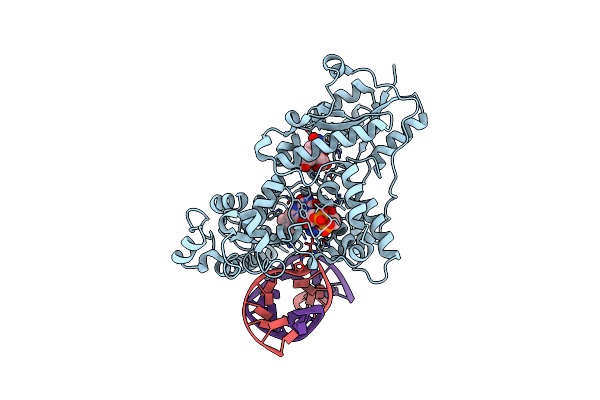 |
Drosophila Melanogaster (6-4) Photolyase H365N Mutant Bound To Ds Dna With A T-T (6-4) Photolesion And Cofactor F0
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-10-13 Classification: Lyase/DNA Ligands: FAD, FO1 |
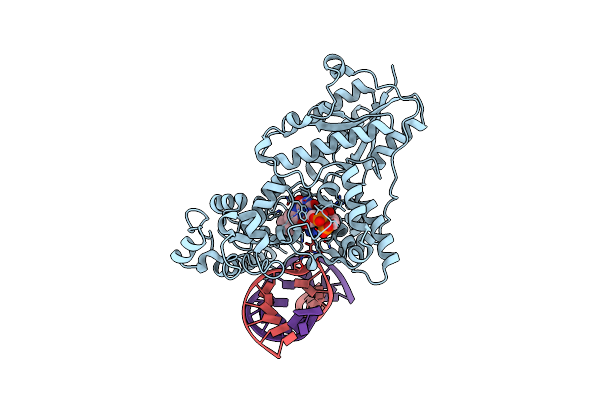 |
Drosophila Melanogaster (6-4) Photolyase H369M Mutant Bound To Ds Dna With A T-T (6-4) Photolesion
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2009-10-13 Classification: Lyase/DNA Ligands: FAD |
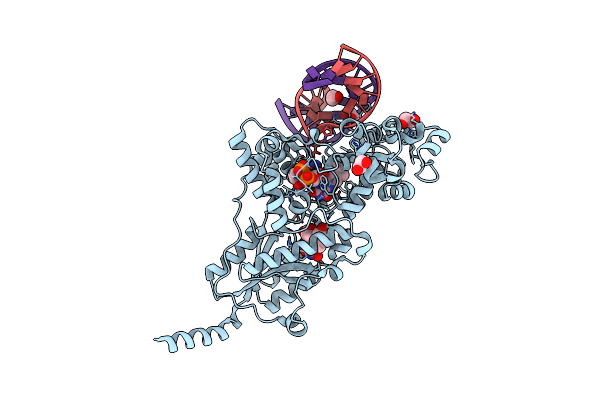 |
Drosophila Melanogaster (6-4) Photolyase Bound To Ds Dna With A T-T (6-4) Photolesion And F0 Cofactor
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-06-16 Classification: Lyase/DNA Ligands: EDO, FAD, FO1 |
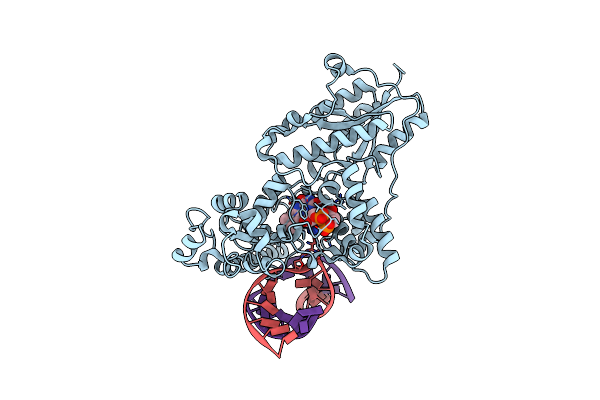 |
Drosophila Melanogaster (6-4) Photolyase Bound To Double Stranded Dna Containing A T(6-4)C Photolesion
Organism: Drosophila melanogaster, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2009-03-17 Classification: LYASE/DNA Ligands: FAD |
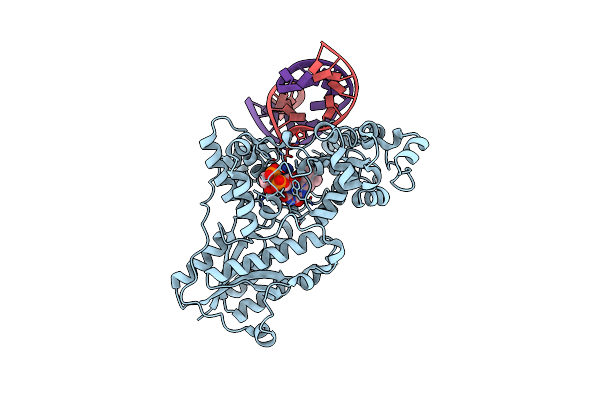 |
Drosophila Melanogaster (6-4) Photolyase Bound To Ds Dna With A T-T (6-4) Photolesion
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-04 Classification: Lyase/DNA Ligands: FAD |
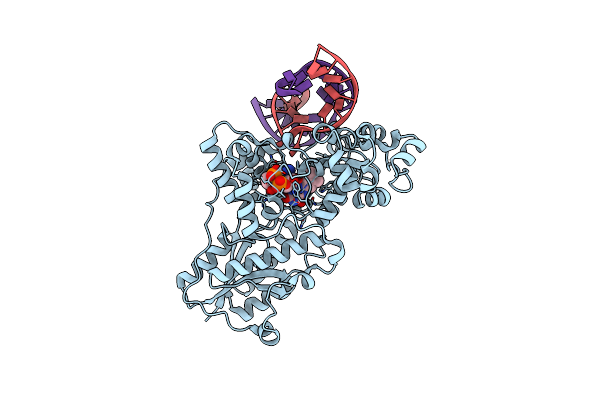 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-11-04 Classification: Lyase/DNA Ligands: FAD |

