Search Count: 96
 |
Organism: Nematostella vectensis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2021-07-07 Classification: ANTIVIRAL PROTEIN Ligands: SF4, SAM, CTP, NA |
 |
Organism: Hypocrea virens (strain gv29-8 / fgsc 10586)
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2021-06-30 Classification: ANTIVIRAL PROTEIN Ligands: SAM, SF4, UTP |
 |
Organism: Methylobacterium extorquens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-02-14 Classification: OXIDOREDUCTASE Ligands: FES, SF4 |
 |
Crystal Structure Of A 5'-Methylthioadenosine/S-Adenosylhomocysteine (Mta/Sah) Nucleosidase (Mtan) From Colwellia Psychrerythraea 34H (Cps_4743, Target Psi-029300) In Complex With Adenine At 2.27 A Resolution
Organism: Colwellia psychrerythraea
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: GLY, ADE |
 |
Crystal Structure Of Stachydrine Demethylase With N-Methyl Proline From Low X-Ray Dose Composite Datasets
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: FES, FE, 3BY, TRS, GOL, NCO |
 |
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: FES, FE, GOL, NCO, AZI |
 |
Crystal Structure Of Stachydrine Demethylase In Complex With Cyanide, Oxygen, And N-Methyl Proline In A New Orientation
Organism: Sinorhizobium meliloti 1021
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2015-07-15 Classification: OXIDOREDUCTASE Ligands: FES, FE, TRS, NCO, 3BY, OXY, CYN |
 |
Crystal Structure Of Homoserine Kinase From Yersinia Pestis Nepal516, Nysgrc Target 032715
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-11-12 Classification: TRANSFERASE Ligands: CIT |
 |
Crystal Structure Of A Putative Pyrimidine-Specific Ribonucleoside Hydrolase (Riha) Protein From Shewanella Loihica Pv-4 (Shew_0697, Target Psi-029635) With Divalent Cation And Peg 400 Bound At The Active Site
Organism: Shewanella loihica
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-11-12 Classification: HYDROLASE Ligands: CA, 1PE |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Streptococcus Agalactiae 2603V/R, Nysgrc Target 030935
Organism: Streptococcus agalactiae lmg 15084
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-07-23 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of Hypoxanthine Phosphoribosyltransferase From Brachybacterium Faecium Dsm 4810, Nysgrc Target 029763.
Organism: Brachybacterium faecium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-07-02 Classification: TRANSFERASE Ligands: MG |
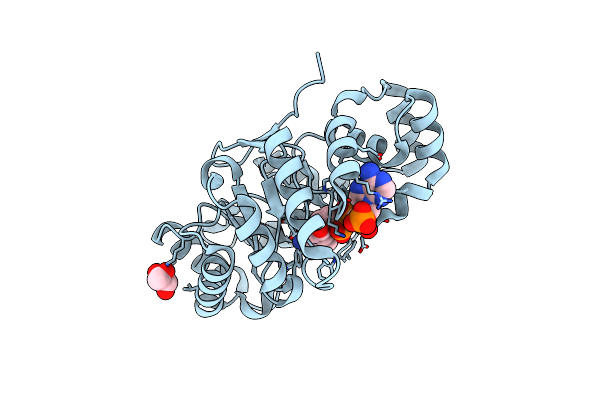 |
Crystal Structure Of A Putative Oxidoreductase From Sinorhizobium Meliloti 1021 In Complex With Nadp
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: NAP, GOL |
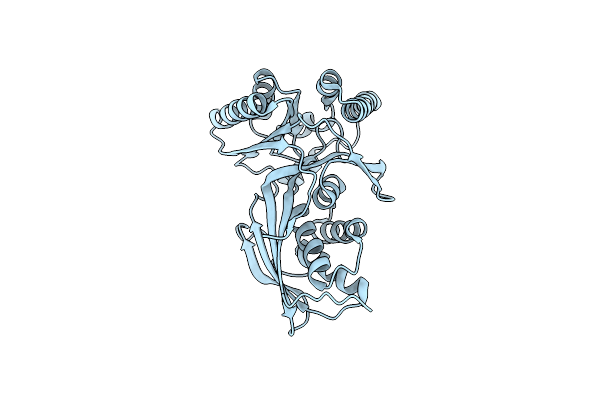 |
Crystal Structure Of Homoserine Kinase From Cytophaga Hutchinsonii Atcc 33406, Nysgrc Target 032717.
Organism: Cytophaga hutchinsonii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-04-02 Classification: TRANSFERASE |
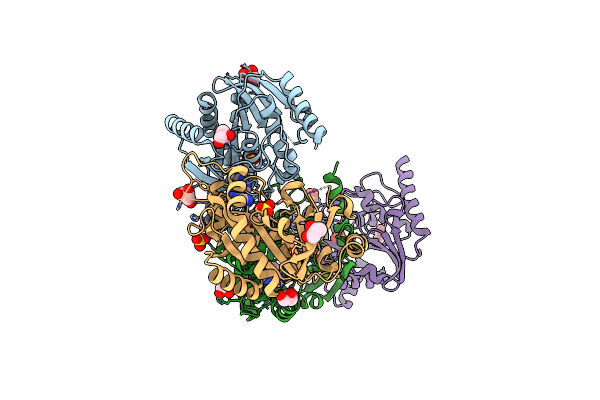 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Porphyromonas Gingivalis Atcc 33277, Nysgrc Target 30972
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: D5M, SO4, GOL |
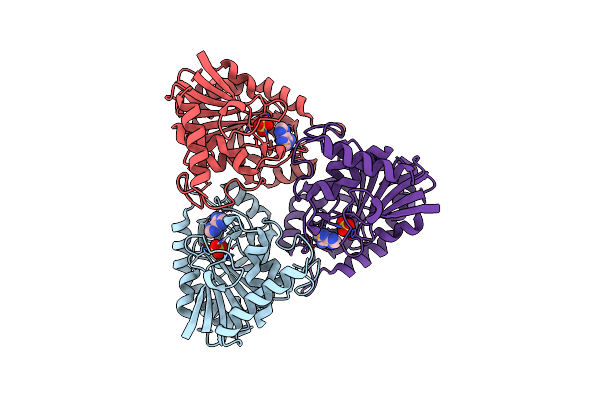 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Porphyromonas Gingivalis Atcc 33277, Nysgrc Target 030972, Orthorhombic Symmetry
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: ADE, SO4 |
 |
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase Family Protein From Klebsiella Pneumoniae 342
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: PE4, CIT |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Meiothermus Ruber Dsm 1279 Complexed With Sulfate.
Organism: Meiothermus ruber
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2013-10-09 Classification: TRANSFERASE Ligands: MG, SO4 |
 |
Crystal Structure Of Uridine Phosphorylase From Vibrio Fischeri Es114 Complexed With 6-Hydroxy-1-Naphthoic Acid, Nysgrc Target 029520.
Organism: Vibrio fischeri
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: SO4, 61N, DMS |
 |
Crystal Structure Of Uridine Phosphorylase From Vibrio Fischeri Es114 Complexed With Dmso, Nysgrc Target 029520.
Organism: Vibrio fischeri
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-09-04 Classification: TRANSFERASE Ligands: SO4, DMS |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Meiothermus Ruber Dsm 1279, Nysgrc Target 029804.
Organism: Meiothermus ruber dsm 1279
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: PO4, MG |

