Search Count: 13
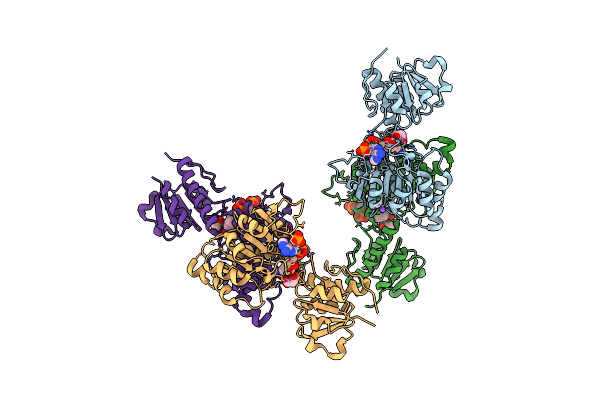 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And Malonate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, MLA, NA |
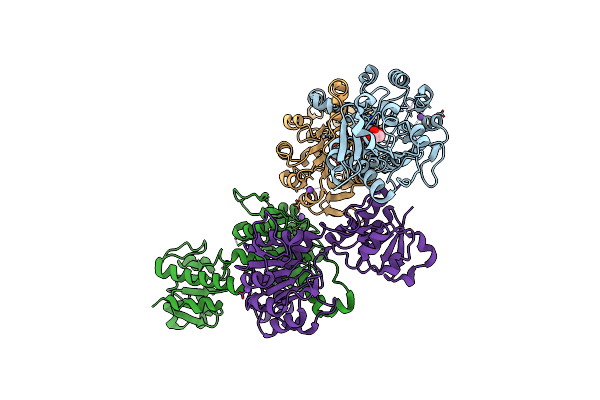 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Citrate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE |
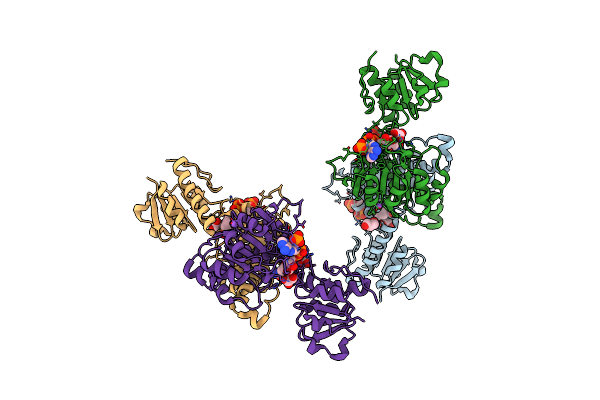 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: OXL, NDP, GOL, CL, NA |
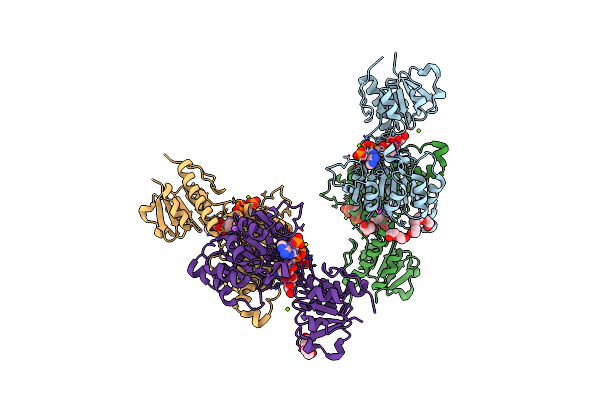 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And 2-Keto-D-Gluconic Acid
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, 8YV, NA, PEG, CL, PG4, MG, GOL |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Apo Form
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: GOL, CL |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Apo Form
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Thermoplasma acidophilum
Method: ELECTRON MICROSCOPY Resolution:7.00 Å Release Date: 2016-07-27 Classification: HYDROLASE |
 |
Organism: Thermoplasma acidophilum
Method: ELECTRON MICROSCOPY Resolution:7.80 Å Release Date: 2016-07-27 Classification: HYDROLASE |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With 2'-Phospho-Adp-Ribose
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: ACT, CL, A2R |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-08 Classification: OXIDOREDUCTASE Ligands: CL, OXD, EPE, GOL, NDP |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Complex With Nadp And Sulfate
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-24 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, PEG, GOL, CL |
 |
Crystal Structure Of The Apbe Protein (Tm1553) From Thermotoga Maritima Msb8 At 1.58 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2005-03-22 Classification: BIOSYNTHETIC PROTEIN Ligands: UNL, MRD, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2004-06-22 Classification: DNA BINDING PROTEIN |

