Search Count: 16
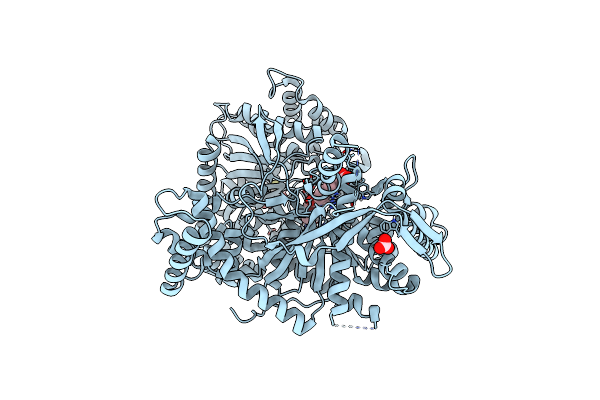 |
A Glucose-Based Molecular Rotor Probes The Catalytic Site Of Glycogen Phosphorylase.
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: I0F, CO3, BME |
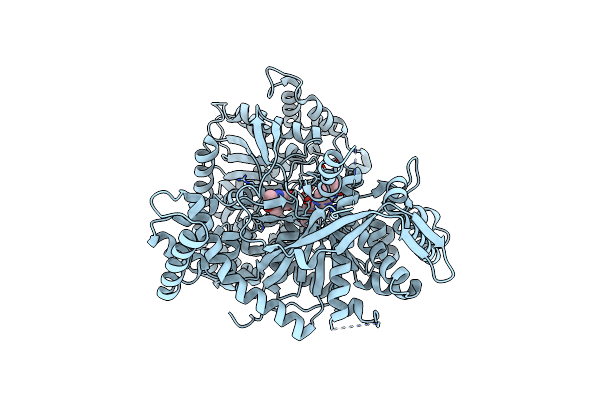 |
A Potent Fluorescent Inhibitor Of Glycogen Phosphorylase As A Catalytic Site Probe.
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2017-11-29 Classification: FLUORESCENT PROTEIN Ligands: PLP, 7LS |
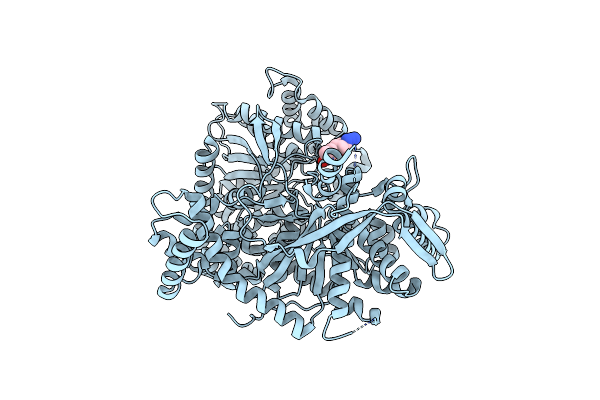 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: AZZ |
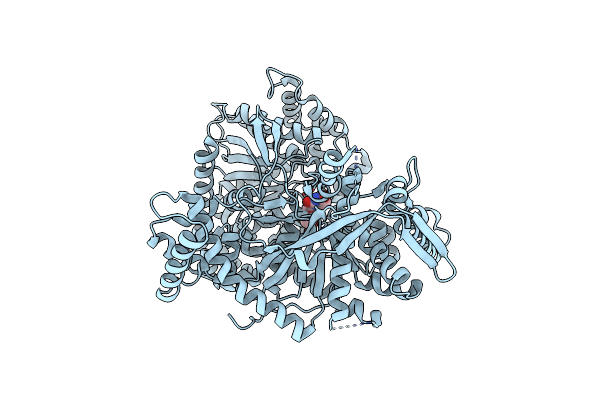 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: CJB |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: THM |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: RDD |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: PLP, CKB |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: PLP, C3B |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-11-18 Classification: TRANSFERASE Ligands: C4B |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-06-03 Classification: TRANSFERASE, APOPTOSIS Ligands: 0AS |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2008-06-03 Classification: TRANSFERASE, APOPTOSIS Ligands: 0MA |
 |
The Crystal Structure Of Glycogen Phosphorylase B In Complex With A Potent Allosteric Inhibitor
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-08-07 Classification: TRANSFERASE Ligands: OFF |
 |
Crystal Structure Of The Glycogen Phosphorylase B / N-(Beta-D-Glucopyranosyl)Oxamic Acid Complex
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2006-05-09 Classification: TRANSFERASE Ligands: PLP, 4GP |
 |
Crystal Structure Of The Glycogen Phosphorylase B / Methyl-N-(Beta-D-Glucopyranosyl)Oxamate Complex
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2006-05-09 Classification: TRANSFERASE Ligands: PLP, 6GP |
 |
Crystal Structure Of The Glycogen Phosphorylase B / Ethyl-N-(Beta-D-Glucopyranosyl)Oxamate Complex
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2006-05-09 Classification: TRANSFERASE Ligands: PLP, 7GP |
 |
Crystal Structure Of The Glycogen Phosphorylase B / N-(Beta-D-Glucopyranosyl)-N'-Cyclopropyl Oxalamide Complex
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2006-05-09 Classification: TRANSFERASE Ligands: PLP, 8GP |

