Search Count: 29
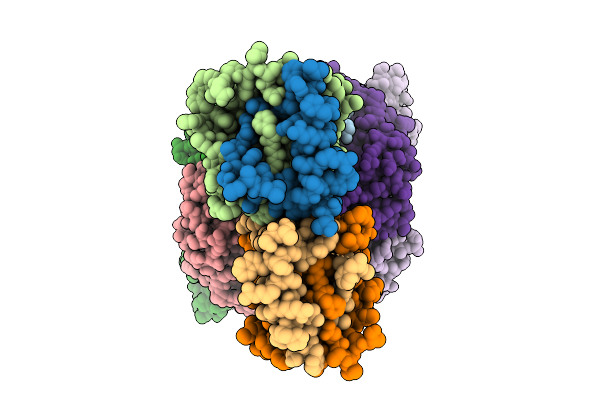 |
Thermus Thermophilus Mrec-Mred Complex With An Internal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
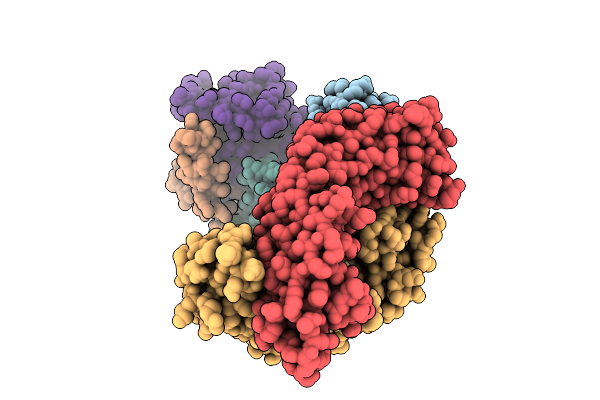 |
Thermus Thermophilus Mrec-Mred Complex With A C-Terminal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
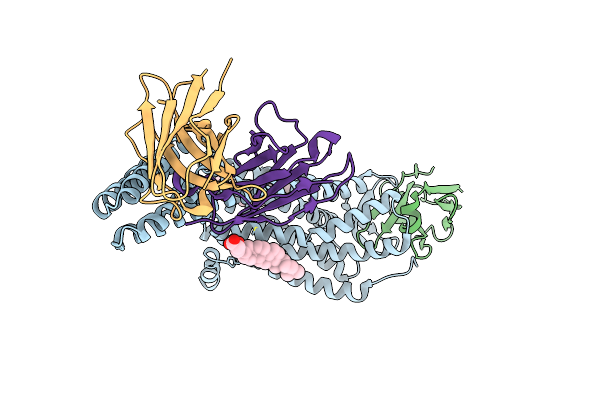 |
Structure Of At118-H Nanobody Antagonist In Complex With The Angiotensin Ii Type I Receptor
Organism: Camelidae, Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
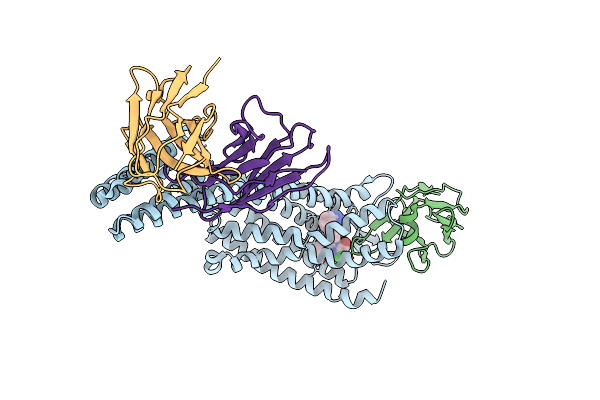 |
Structure Of At118-L Nanobody Antagonist In Complex With The Angiotensin Ii Type I Receptor And Losartan
Organism: Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: LSN |
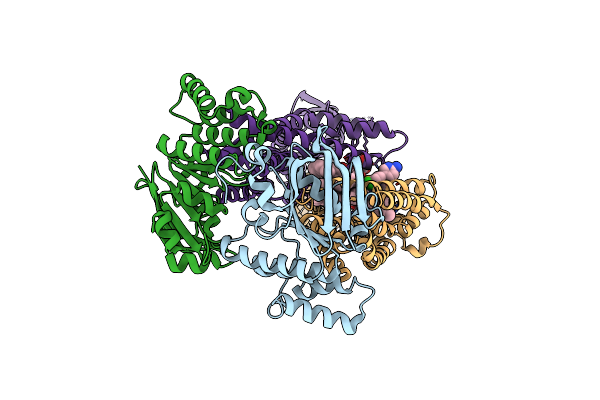 |
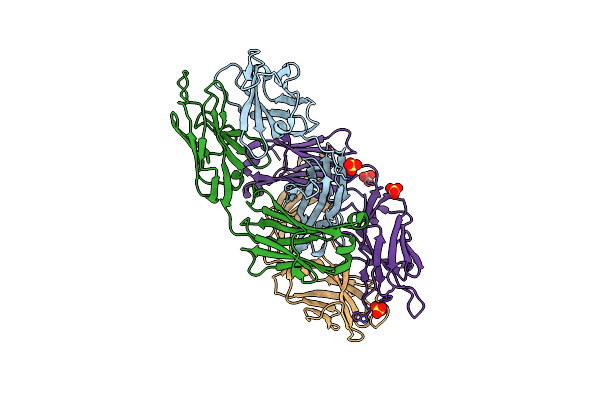
Acinetobacter Baylyi Lptb2Fg Bound To Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: Y75, JSG, LMT |
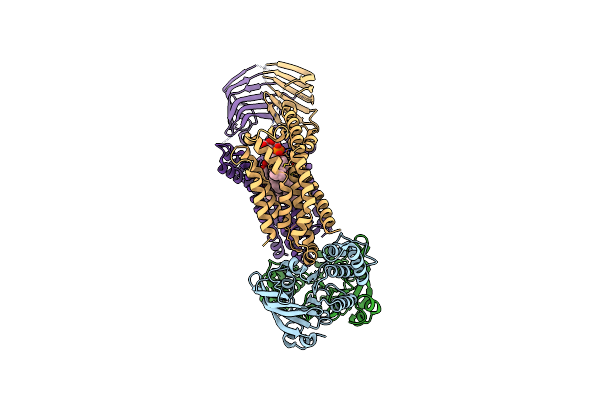 |
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG |
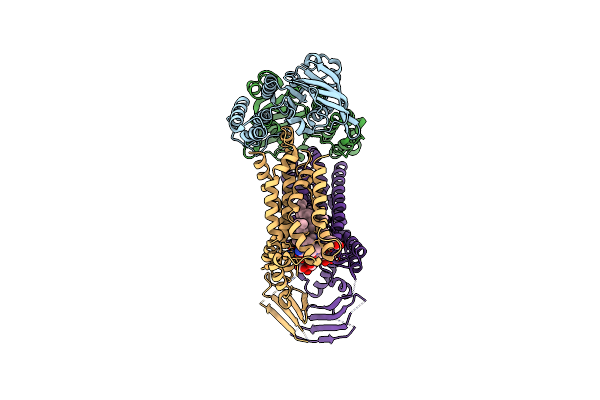 |
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG, VB6, LMT |
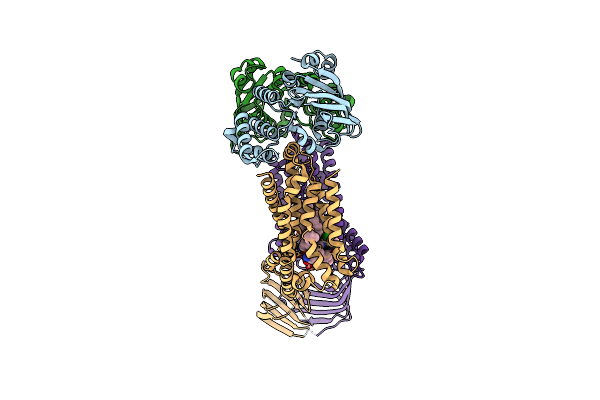 |
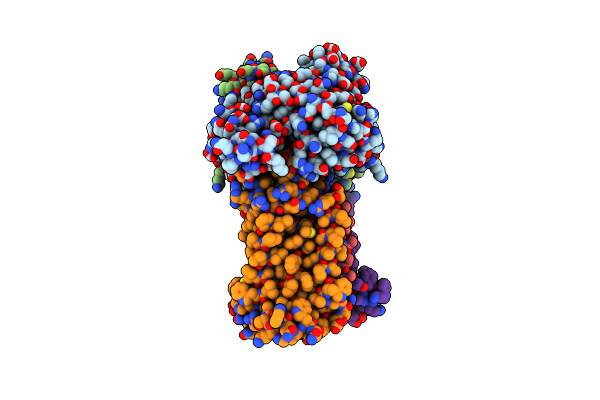
Acinetobacter Baylyi Lptb2Fg Bound To Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG, MG3 |
 |
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT |
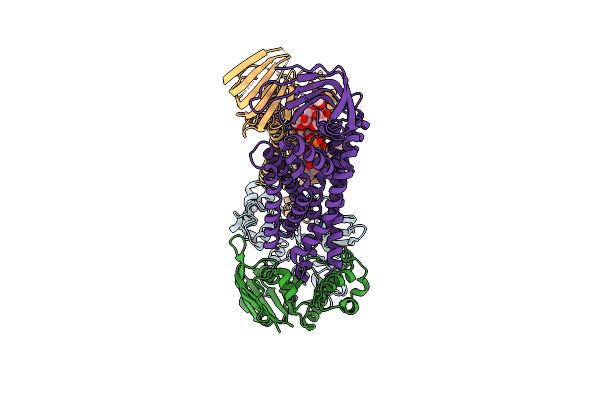 |
Acinetobacter Baylyi Lptb2Fg Bound To Acinetobacter Baylyi Lipopolysaccharide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: LMT, WJR |
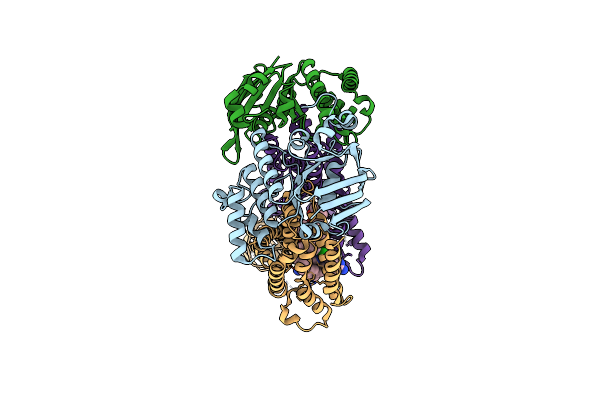 |
Acinetobacter Baylyi Lptb2Fg Bound To Acinetobacter Baylyi Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: WJW, Y75 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-06-16 Classification: IMMUNE SYSTEM Ligands: SO4 |
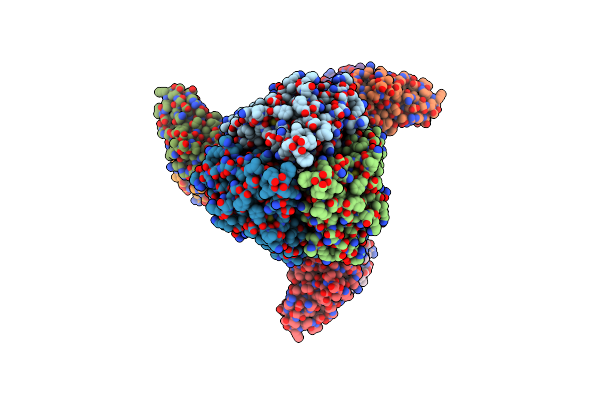 |
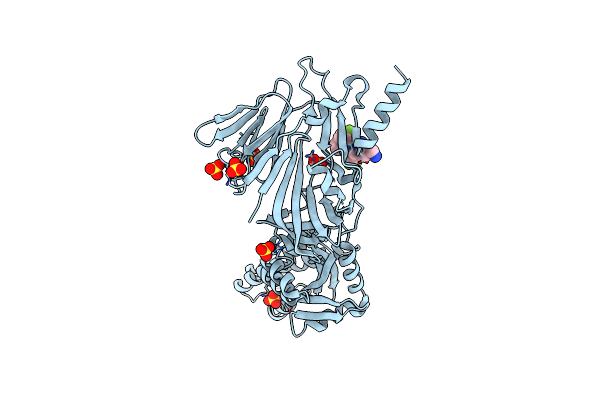
Prefusion Rsv F Glycoprotein Bound By Neutralizing Site V-Directed Antibody Adi-14442
Organism: Respiratory syncytial virus a2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-06-16 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
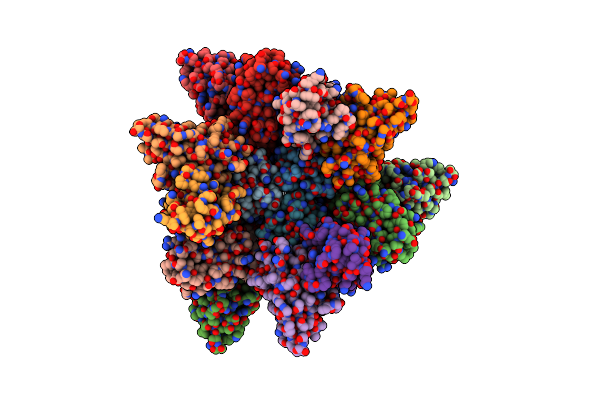
Organism: Respiratory syncytial virus
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2021-04-21 Classification: VIRAL PROTEIN Ligands: WVA, SO4 |
 |
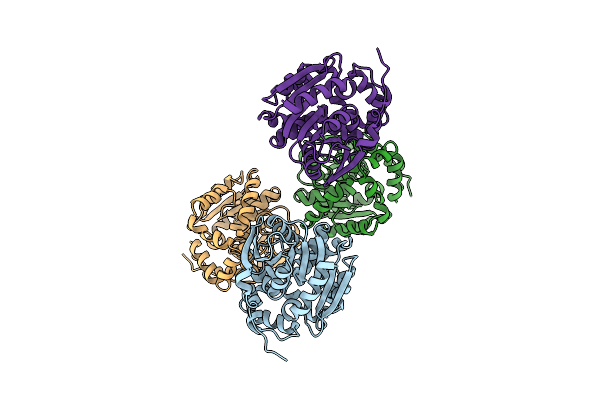
Organism: Respiratory syncytial virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-04-21 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-17 Classification: HYDROLASE |
 |
Organism: Burkholderia cenocepacia (strain atcc baa-245 / dsm 16553 / lmg 16656 / nctc 13227 / j2315 / cf5610)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-03-17 Classification: HYDROLASE |
 |
Organism: Burkholderia cenocepacia (strain mc0-3)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-03-17 Classification: HYDROLASE |
 |
Organism: Zaire ebolavirus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structural Coordination Of Polymerization And Crosslinking By A Peptidoglycan Synthase Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2020-03-18 Classification: MEMBRANE PROTEIN |

