Search Count: 31
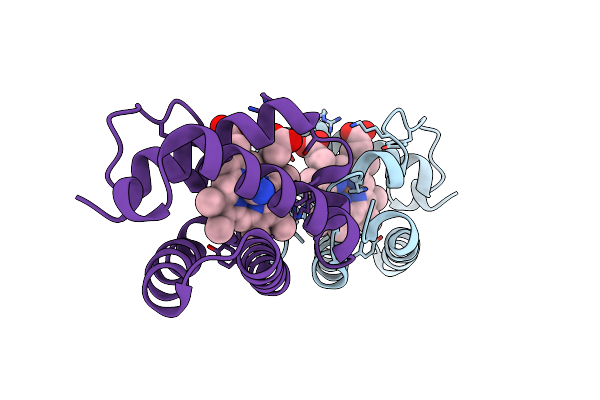 |
Crystal Structure Of Shewanella Benthica Group 1 Truncated Hemoglobin C51S C71S Y108A Variant
Organism: Shewanella benthica kt99
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-03 Classification: HEME BINDING PROTEIN Ligands: HEM, CYN |
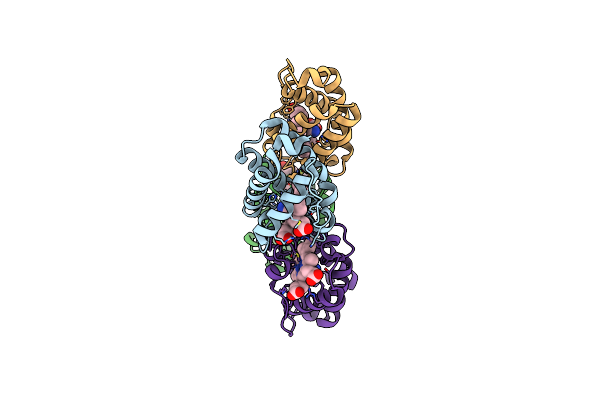 |
Crystal Structure Of Shewanella Benthica Group 1 Truncated Hemoglobin L80A C51S C71S Variant
Organism: Shewanella benthica kt99
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-04-03 Classification: HEME-BINDING PROTEIN Ligands: HEM, CYN |
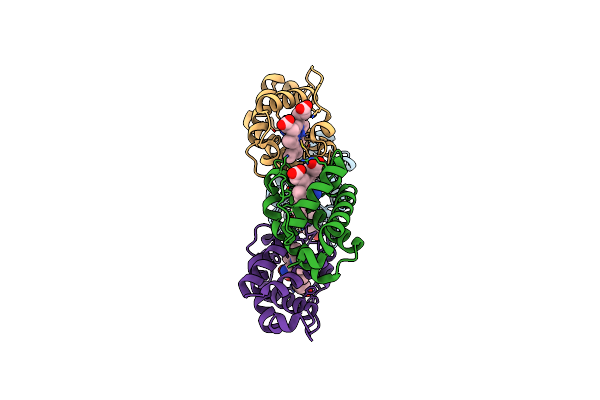 |
Crystal Structure Of Shewanella Benthica Group 1 Truncated Hemoglobin Y34F C51S C71S Variant (Cyanomet)
Organism: Shewanella benthica kt99
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-04-03 Classification: HEME-BINDING PROTEIN Ligands: HEM, CYN |
 |
Organism: Pseudomonas phage vb_paem_e217
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN |
 |
Organism: Pseudomonas phage vb_paem_e217
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-09-07 Classification: HYDROLASE Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-06-22 Classification: PROTEIN TRANSPORT |
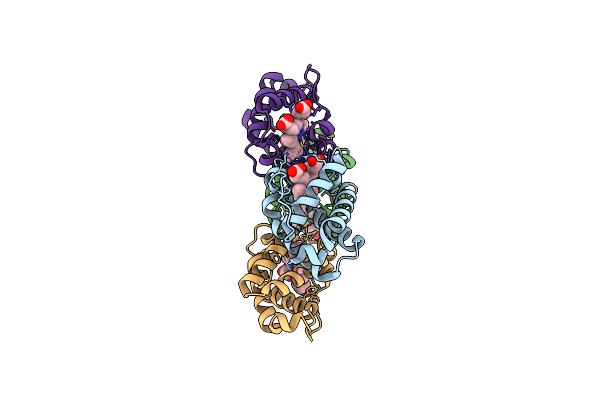 |
Crystal Structure Of Shewanella Benthica Group 1 Truncated Hemoglobin C51S C71S Y34F Variant
Organism: Shewanella benthica kt99
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-02-16 Classification: HEME BINDING PROTEIN Ligands: HEM |
 |
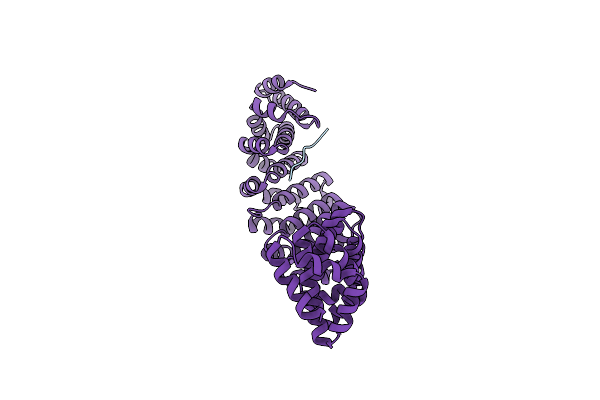
Structure Of The M298F Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Axial Ligand.
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE Ligands: CU |
 |
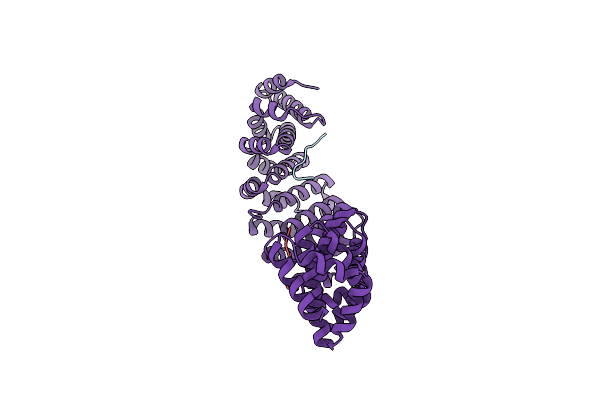
Structure Of The M298L Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Axial Ligand
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-02-09 Classification: OXIDOREDUCTASE Ligands: CU |
 |
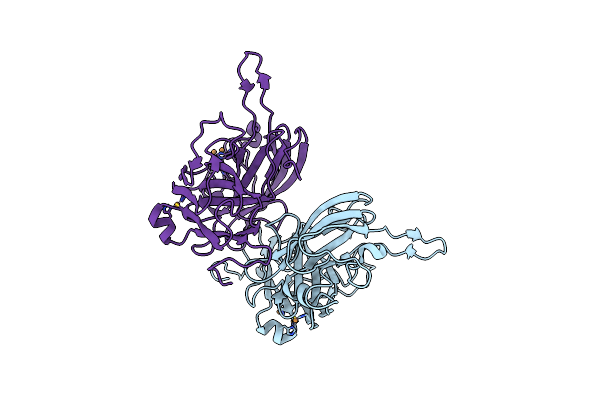
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2022-01-19 Classification: PROTEIN TRANSPORT |
 |
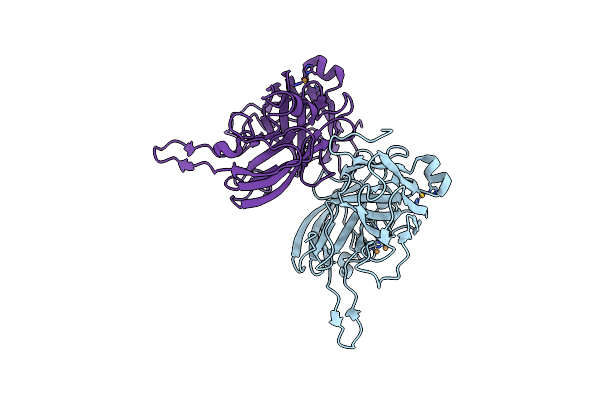
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-01-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2022-01-19 Classification: PROTEIN TRANSPORT Ligands: HOH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-01-19 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-01-19 Classification: PROTEIN TRANSPORT |
 |
Structure Of The Streptomyces Coelicolor Small Laccase - Cubic Crystal Form
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Structure Of The M198F M298F Double Mutant Of The Streptomyces Coelicolor Small Laccase T1 Copper Site
Organism: Streptomyces coelicolor (strain atcc baa-471 / a3(2) / m145)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PG4, CU |
 |
Organism: Pseudomonas virus pap3
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2020-11-11 Classification: DNA BINDING PROTEIN, VIRAL PROTEIN |
 |
Organism: Pseudomonas phage nv1
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2020-11-11 Classification: VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2017-09-13 Classification: PROTEIN TRANSPORT / NUCLEAR PROTEIN |

