Search Count: 19
 |
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
 |
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-2-Tetralone
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDH, EDO |
 |
Imine Reductase Ir91 From Kribbella Flavida With Nadp+ And 5-Methoxy-(S)-2-(N-Methylamino)Tetralin
Organism: Kribbella flavida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, NAP, A1JDG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: GOL, VTY |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-08-07 Classification: HYDROLASE Ligands: IMD, EDO |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-07-31 Classification: HYDROLASE |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: MPD |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-07-31 Classification: HYDROLASE |
 |
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With 3D Fragment 2548
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-02-21 Classification: CARBOHYDRATE Ligands: EDO, UE0, SO4 |
 |
Organism: Ensifer adhaerens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-03-08 Classification: OXIDOREDUCTASE Ligands: NAP, K |
 |
Organism: Ensifer adhaerens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-03-08 Classification: OXIDOREDUCTASE Ligands: NAP, IOD |
 |
Organism: Micromonospora sp. rc5
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
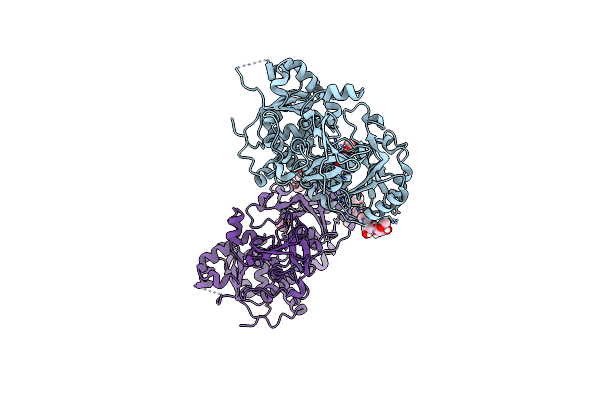
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, SO4, NAG |
 |
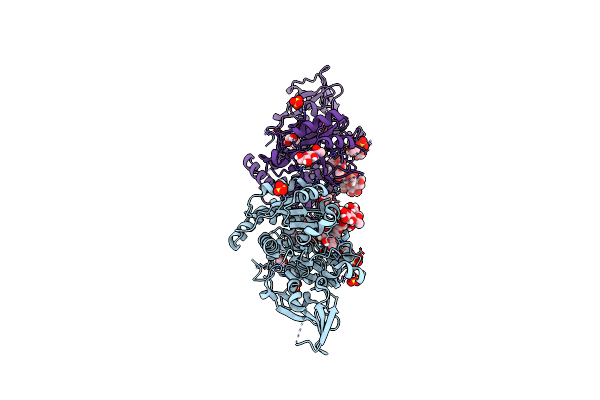
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: NAG, EDO |
 |
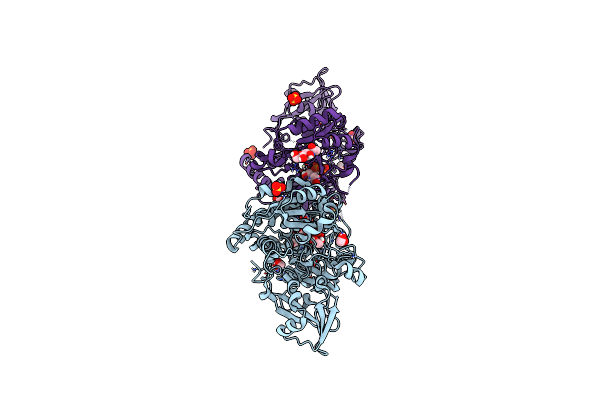
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, NAG, SO4 |
 |
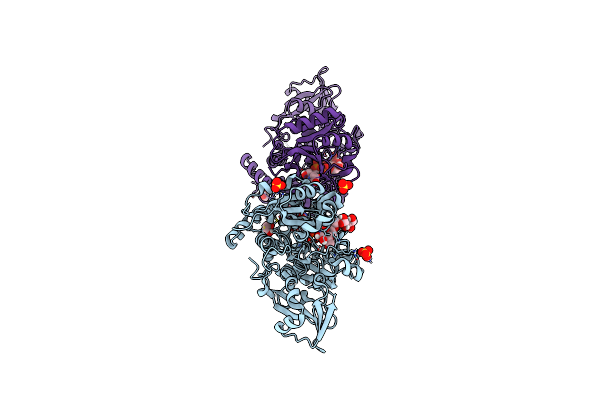
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: UDP, EDO, NAG, SO4 |
 |
Crystal Structure Of Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp And Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: UDP, EDO, SO4 |
 |
Crystal Structure Of Unliganded Mgat5 (Alpha-1,6-Mannosylglycoprotein 6-Beta-N-Acetylglucosaminyltransferase V) Luminal Domain With A Lys329-Ile345 Loop Truncation, In Complex With Udp-2-Deoxy-2-Fluoroglucose And Biantennary Pentasaccharide M592
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: EDO, U2F, SO4 |

