Search Count: 22
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2005-10-04 Classification: HYDROLASE Ligands: ZN, CA, IOY |
 |
Crystal Structure Of Aeromonas Proteolytica Aminopeptidase In Complex With Bestatin
Organism: Vibrio proteolyticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-09-27 Classification: HYDROLASE Ligands: ZN, BES |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2005-06-14 Classification: HYDROLASE Ligands: ZN, CA, DTR |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2005-06-14 Classification: HYDROLASE Ligands: ZN, CA, DPN |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2005-06-14 Classification: HYDROLASE Ligands: ZN, CA, MED |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-05-31 Classification: HYDROLASE Ligands: ZN, CA, TRP |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2005-05-31 Classification: HYDROLASE Ligands: ZN, CA, PHI |
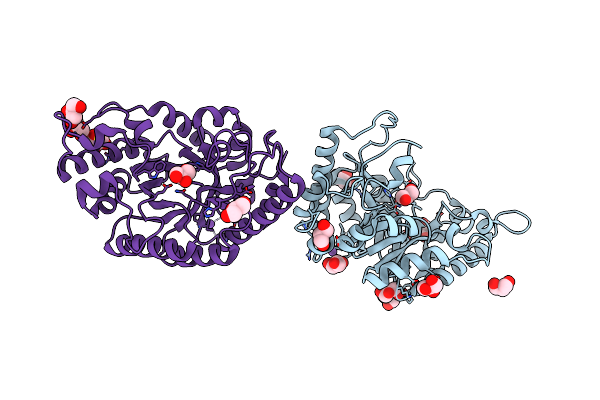 |
The High-Resolution Crystal Structure Of Ixt6, A Thermophilic, Intracellular Xylanase From G. Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2003-11-25 Classification: HYDROLASE |
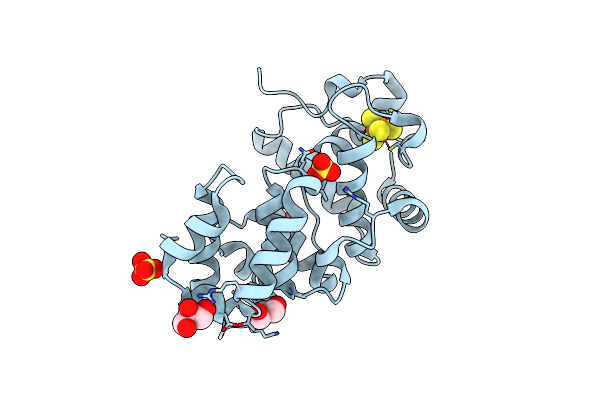 |
Crystal Structure Of The Core Fragment Of Muty From E.Coli At 1.2A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
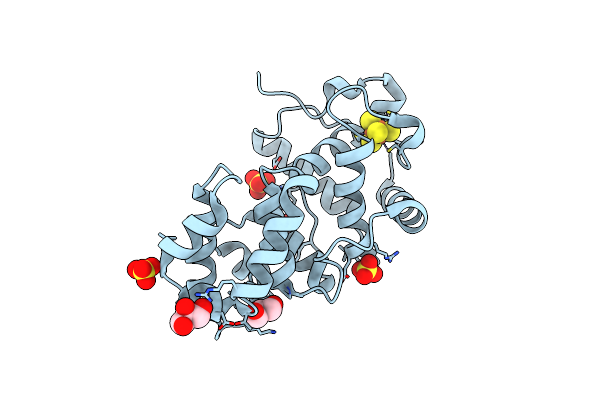 |
Crystal Structure Of The Core Fragment Of Muty From E.Coli At 1.55A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
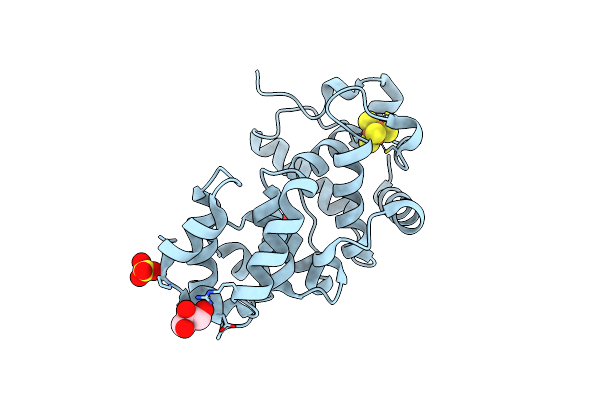 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |
 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4 |
 |
Crystal Structure Of A Trapped Reaction Intermediate Of The Dna Repair Enzyme Endonuclease Viii With Brominated-Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2002-10-04 Classification: HYDROLASE/DNA Ligands: ZN, SO4, GOL |
 |
Crystal Structure Of E.Coli Formamidopyrimidine-Dna Glycosylase (Fpg) Covalently Trapped With Dna
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2002-06-14 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Organism: Bacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2002-01-04 Classification: GLYCOSIDASE Ligands: GLA, GLC, SO4 |
 |
Crystal Structure Of The Streptomyces Griseus Aminopeptidase Complexed With L-Leucine
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-08-22 Classification: HYDROLASE Ligands: ZN, CA, LEU |
 |
Crystal Structure Of The Streptomyces Griseus Aminopeptidase Complexed With L-Phenylalanine
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-08-22 Classification: HYDROLASE Ligands: ZN, CA, PHE |

