Search Count: 9
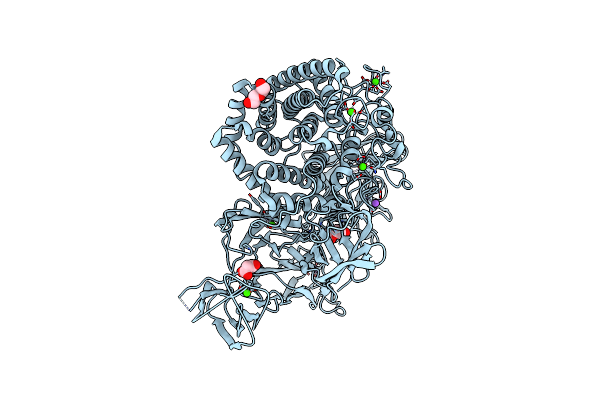 |
Structural Dynamics And Catalytic Properties Of A Multi-Modular Xanthanase, Native.
Organism: Paenibacillus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: CA, NA, CL, PEG, MLI |
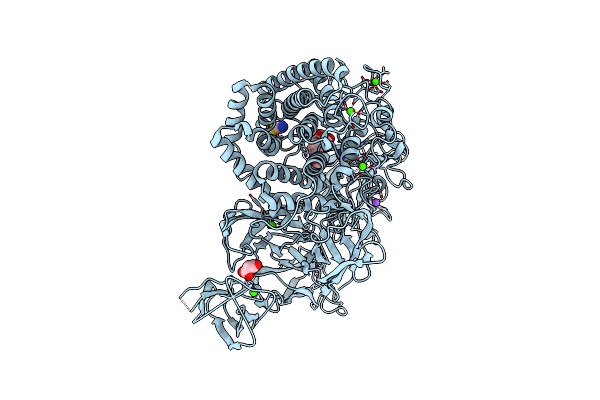 |
Structural Dynamics And Catalytic Properties Of A Multi-Modular Xanthanase (Pt Derivative)
Organism: Paenibacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-08-29 Classification: HYDROLASE Ligands: PT, B3P, SCN, CA, NA, CL, MLI |
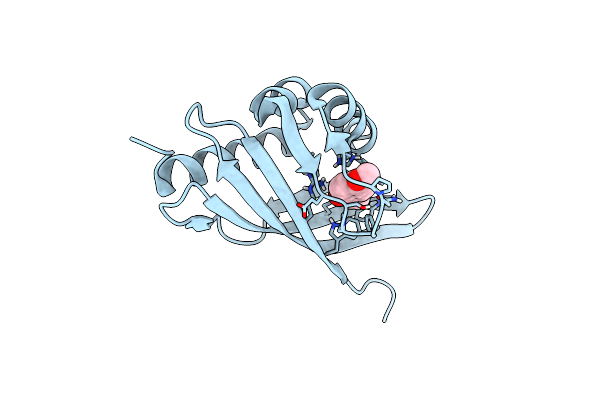 |
Crystal Structure Of Retro-Aldolase Ra110.4-6 Complexed With Inhibitor 1-(6-Methoxy-2-Naphthalenyl)-1,3-Butanedione
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-12 Classification: ISOMERASE Ligands: LLK |
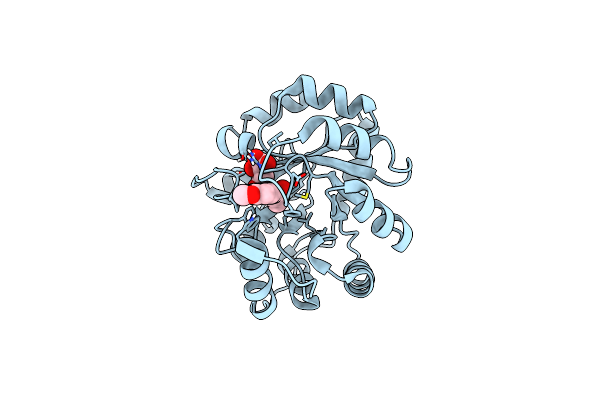 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-07 Classification: DE NOVO PROTEIN Ligands: 3NK, MLT |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2012-11-07 Classification: LYASE Ligands: 3NK |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-11-07 Classification: LYASE Ligands: 3NK |
 |
Structural Analyses Of Covalent Enzyme-Substrate Analogue Complexes Reveal Strengths And Limitations Of De Novo Enzyme Design
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-11-23 Classification: LYASE/LYASE INHIBITOR Ligands: 3NK, SO4 |
 |
Robust Computational Design, Optimization, And Structural Characterization Of Retroaldol Enzymes
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-06-29 Classification: LYASE Ligands: SO4 |
 |
Robust Computational Design, Optimization, And Structural Characterization Of Retroaldol Enzymes
Organism: Artificial gene
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2011-06-29 Classification: LYASE Ligands: SO4 |

