Search Count: 18
 |
Dihydrolipoamide Acetyltransferase (E2) Psbd In Complex With The Pyruvate Dehydrogenase (E1) Binding Domain From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PROTEIN BINDING |
 |
Dihydrolipoyl Dehydrogenase (E3) In Complex With The Binding Domain Of Dihydrolipoamide Acetyltransferase (E2) From The E. Coli Pyruvate Dehydrogenase Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: FLAVOPROTEIN Ligands: FAD |
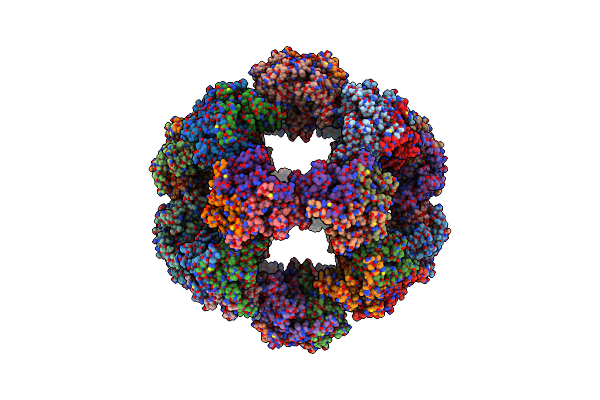 |
60-Meric Complex Of Dihydrolipoamide Acetyltransferase (E2) Of The Human Pyruvate Dehydrogenase Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: STRUCTURAL PROTEIN |
 |
2.5 A Cryo-Em Structure Of The In Vitro Fimd-Catalyzed Assembly Of Type 1 Pilus Rod
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: STRUCTURAL PROTEIN |
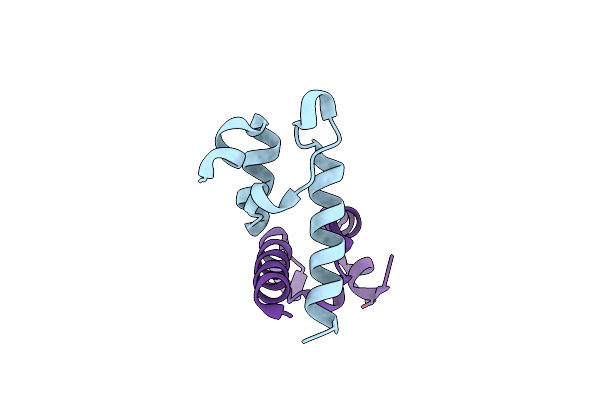 |
Peripheral Subunit Binding Domain Of The E. Coli Dihydrolipoamide Acetyltransferase (E2) Of The Pyruvate Dehydrogenase Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-02-07 Classification: PROTEIN BINDING Ligands: ZN |
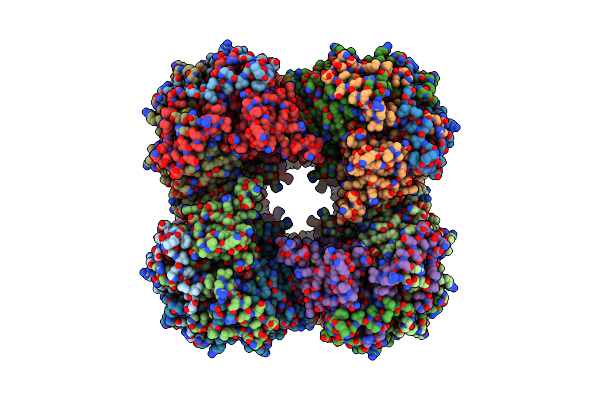 |
24-Meric Catalytic Domain Of Dihydrolipoamide Acetyltransferase (E2) Of The E. Coli Pyruvate Dehydrogenase Complex.
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: TRANSFERASE |
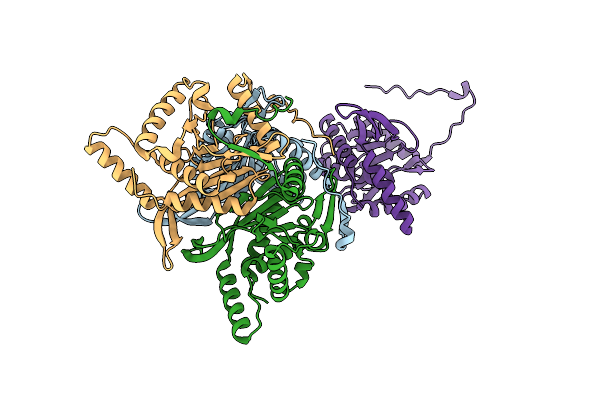 |
Trimeric Catalytic Domain Of The E. Coli Dihydrolipoamide Acetyltransferase (E2) Of The Pyruvate Dehydrogenase Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-02-07 Classification: TRANSFERASE |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-03-29 Classification: DNA BINDING PROTEIN Ligands: ACT, PEG, SO4, EDO |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-03-29 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of The E. Coli Type 1 Pilus Assembly Inhibitor Fimi Bound To Fimc
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-12-08 Classification: STRUCTURAL PROTEIN Ligands: EDO, FMT |
 |
Crystal Structure Of The Ternary Complex Of The E. Coli Type 1 Pilus Proteins Fimc, Fimi And The N-Terminal Domain Of Fimd
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-08 Classification: STRUCTURAL PROTEIN Ligands: EDO |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.85 Å Release Date: 2021-03-31 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of The Ternary Complex Between The Type 1 Pilus Proteins Fimc, Fimi And Fima From E. Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-10-07 Classification: STRUCTURAL PROTEIN Ligands: PEG, EDO |
 |
The Cryo-Electron Microscopy Structure Of The Type 1 Chaperone-Usher Pilus Rod
Organism: Escherichia coli j96
Method: ELECTRON MICROSCOPY Release Date: 2017-11-22 Classification: PROTEIN FIBRIL |
 |
Crystal Structure Of The E. Coli Type 1 Pilus Subunit Fimg (Engineered Variant With Substitution Q134E; N-Terminal Fimg Residues 1-12 Truncated) In Complex With The Donor Strand Peptide Dsf_T4R-T6R-D13N
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-07-06 Classification: CELL ADHESION Ligands: CO |
 |
Crystal Structure Of The E. Coli Type 1 Pilus Subunit Fimg (Engineered Variant With Substitution Q134E; N-Terminal Fimg Residues 1-12 Truncated) In Complex With The Donor Strand Peptide Dsf_Sririrgyvr
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2016-07-06 Classification: CELL ADHESION Ligands: CO, EDO |
 |
Crystal Structure Of The E. Coli Type 1 Pilus Subunit Fimg (Engineered Variant With Substitutions Q134E And S138E; N-Terminal Fimg Residues 1-12 Truncated) In Complex With The Donor Strand Peptide Dsf_T4R-T6R-D13N
Organism: Escherichia coli (strain k12), Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-07-06 Classification: CELL ADHESION Ligands: CO, EDO, 1PE |

