Search Count: 55
 |
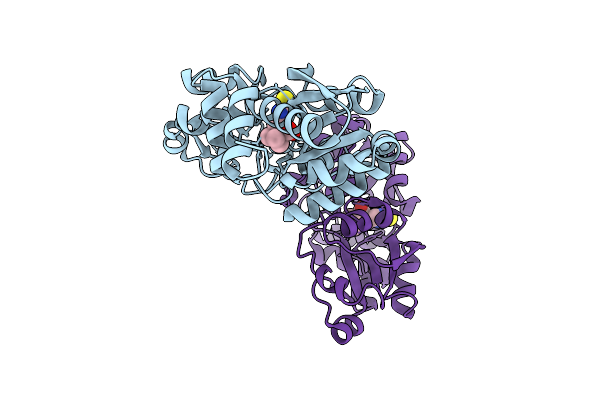
Crystal Structure Of Egtuc Binding Domain Double Mutant I243P T274G Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2026-01-14 Classification: TRANSPORT PROTEIN Ligands: LW8, FLC, CL, NA |
 |
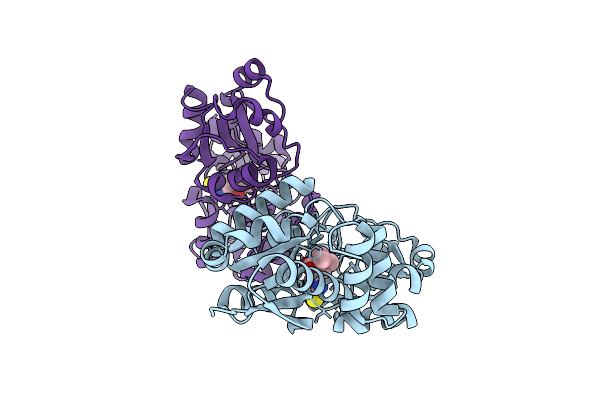
Crystal Structure Of Egtuc Binding Domain Mutant I243A Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8 |
 |
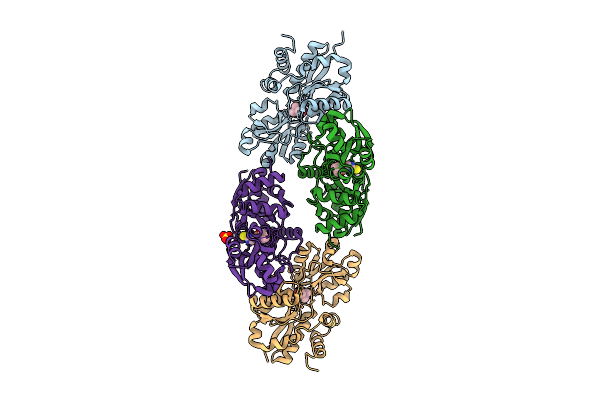
Crystal Structure Of Egtuc Binding Domain Mutant T274G Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8 |
 |
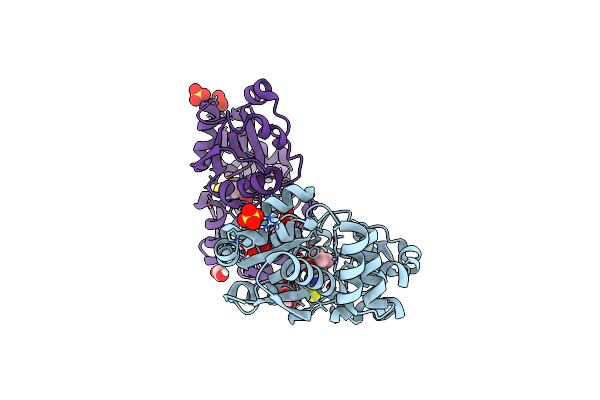
Crystal Structure Of Egtuc Binding Domain Mutant I243P Bound To L-Ergothioneine From S. Pneumoniae
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: LW8, SO4 |
 |
Crystal Structure Of Egtu Solute Binding Domain From Streptococcus Pneumoniae D39 In Complex With L-Ergothioneine
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN Ligands: LW8, 144, SO4, EDO |
 |
Crystal Structure Of Egtu Solute Binding Domain From Streptococcus Pneumoniae D39 In Complex With L-Ergothioneine
Organism: Streptococcus pneumoniae serotype 2 (strain d39 / nctc 7466)
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN Ligands: LW8, EDO |
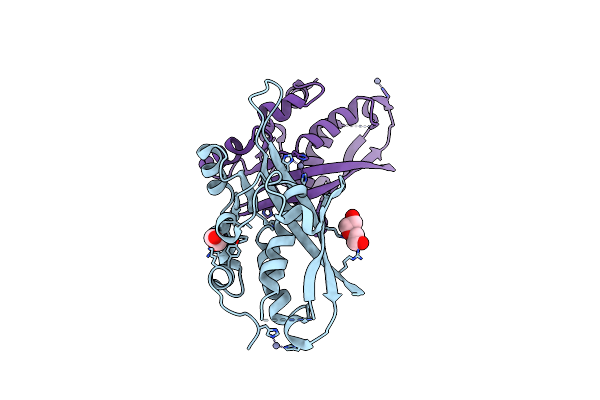 |
Structure Of 6-Carboxy-5,6,7,8-Tetrahydropterin Synthase Paralog Qued2 From Acinetobacter Baumannii
Organism: Acinetobacter baumannii atcc 17978
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-12-07 Classification: LYASE Ligands: EDO, ZN, PEG |
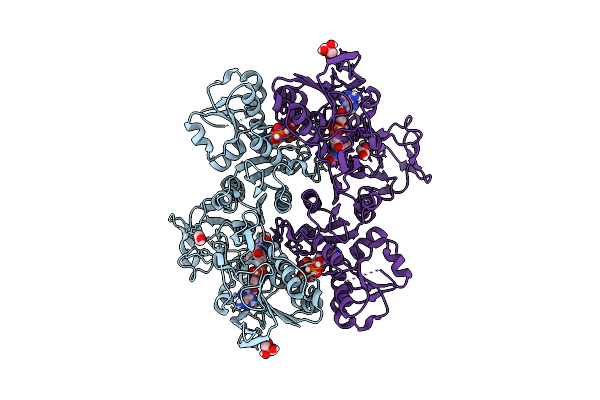 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-09-14 Classification: FLAVOPROTEIN Ligands: FAD, GOL, IOD, EDO, PAP |
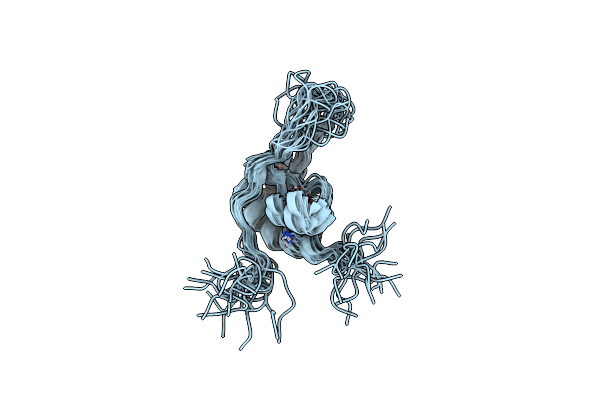 |
Solution Structure Of The Zinc Finger Domain Of Murine Metap1, Complexed With Zng N-Terminal Peptide
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2022-06-01 Classification: METAL BINDING PROTEIN Ligands: ZN |
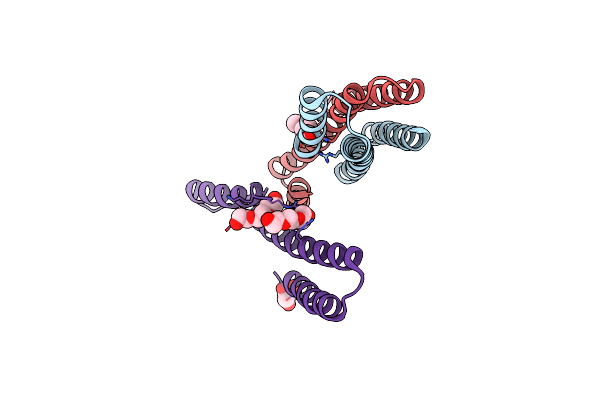 |
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2022-03-09 Classification: TRANSCRIPTION Ligands: CL, PGE, GOL |
 |
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-03-09 Classification: TRANSCRIPTION |
 |
Organism: Streptococcus pneumoniae d39
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-03-09 Classification: TRANSCRIPTION Ligands: MES, NA |
 |
Crystal Structure Of Apo And Reduced Sulfide-Responsive Transcriptional Repressor (Sqrr) From Rhodobacter Capsulatus.
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION Ligands: GOL, SO4 |
 |
Crystal Structure Of C9S Apo And Reduced Sulfide-Responsive Transcriptional Repressor (Sqrr) From Rhodobacter Capsulatus.
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION |
 |
Crystal Structure Of C9S Apo Sulfide-Responsive Transcriptional Repressor (Sqrr) From Rhodobacter Capsulated Bound To Diamide (Tetramethylazodicarboxamide).
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION Ligands: LSY |
 |
Crystal Structure Of C9S Tetrasulfide State Of Sulfide-Responsive Transcriptional Repressor (Sqrr) From Rhodobacter Capsulatus.
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Crystal Structure Of C9S Disulfide State Of Sulfide-Responsive Transcriptional Repressor (Sqrr) From Rhodobacter Capsulatus.
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-01 Classification: TRANSCRIPTION Ligands: SO4, CL |
 |
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With Dodecane-Trimethylamine
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-24 Classification: CELL CYCLE Ligands: CAT |
 |
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With Undecyl-Maltoside
Organism: Streptococcus pneumoniae serotype 2 (strain d39 / nctc 7466)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-24 Classification: CELL CYCLE Ligands: UMQ, SO4, TRS |
 |
Crystal Structure Of Extracellular Domain 1 (Ecd1) Of Ftsx From S. Pneumonie In Complex With N-Decyl-B-D-Maltoside
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2019-04-24 Classification: MEMBRANE PROTEIN Ligands: DMU |

