Search Count: 8
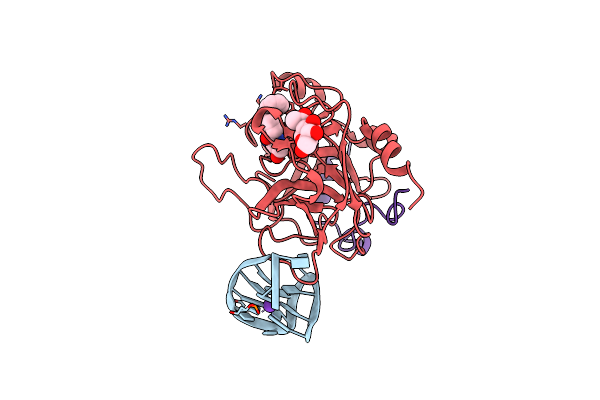 |
X-Ray Structure Of The Complex Between Human Alpha Thrombin And A Modified Thrombin Binding Aptamer (Mtba)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2011-10-19 Classification: HYDROLASE/HYDROLASE INHIBITOR/DNA Ligands: 0G6, NAG, NA, K |
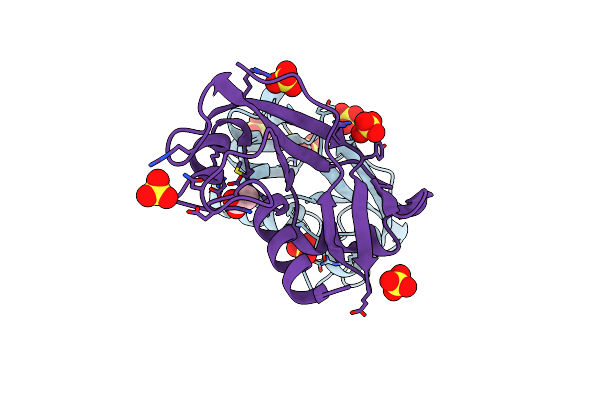 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2010-12-08 Classification: HYDROLASE Ligands: SO4, ACT |
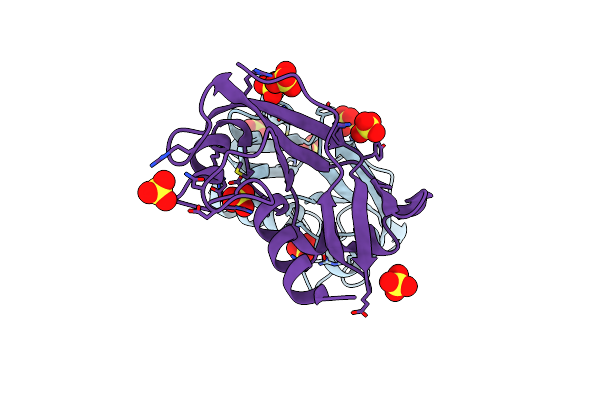 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2010-12-08 Classification: HYDROLASE Ligands: SO4 |
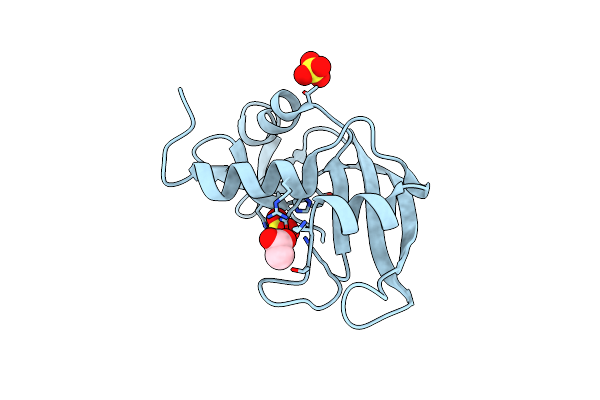 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-11-24 Classification: HYDROLASE Ligands: ACT, SO4 |
 |
Nmr Solution Structure Of The 4:1 Complex Between An Uncharged Distamycin A Analogue And [D(Tggggt)]4
|
 |
 |
 |
Nmr Structure Of A New Modified Thrombin Binding Aptamer Containing A 5'-5' Inversion Of Polarity Site
|

