Search Count: 16
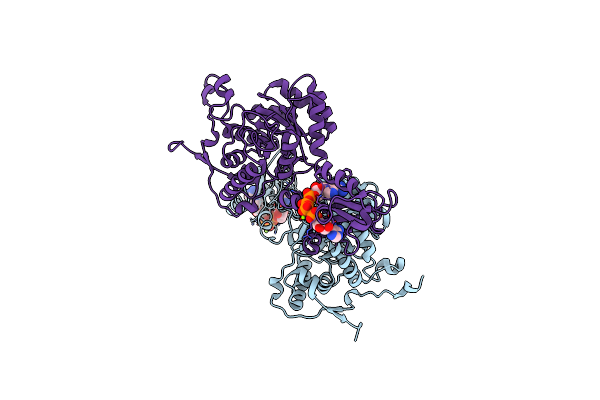 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2022-09-14 Classification: SIGNALING PROTEIN Ligands: B4P, MG |
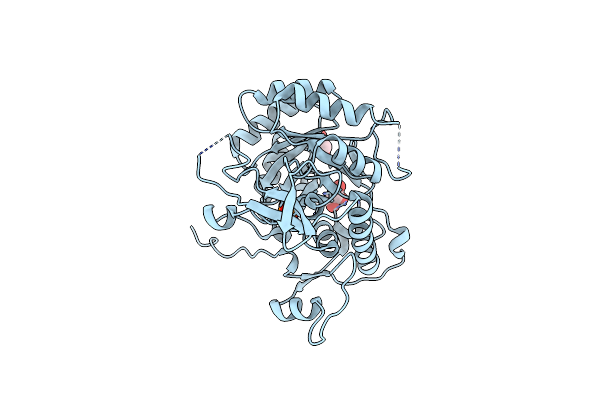 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-09-14 Classification: SIGNALING PROTEIN Ligands: PO4, GOL |
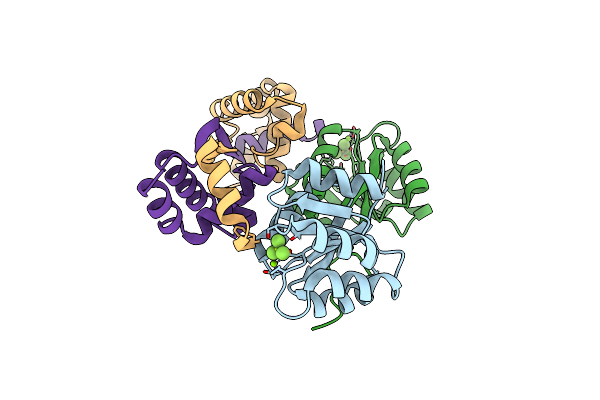 |
Organism: Methanococcus maripaludis x1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-04-27 Classification: METAL BINDING PROTEIN Ligands: MG, BEF |
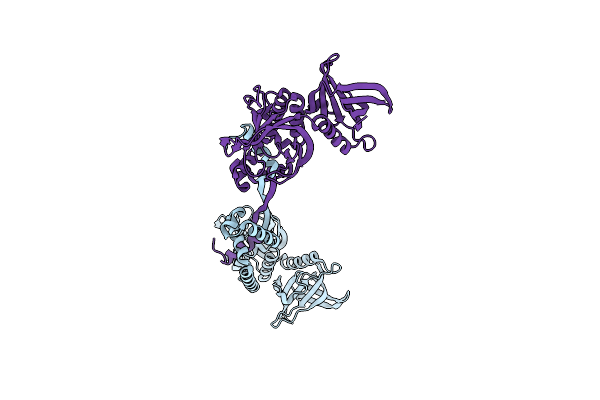 |
Organism: Pyrococcus horikoshii (strain atcc 700860 / dsm 12428 / jcm 9974 / nbrc 100139 / ot-3)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-04-27 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of A Parb E93A Mutant From Myxococcus Xanthus Bound To Cdp And Monothiophosphate
Organism: Myxococcus xanthus (strain dk1622)
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-09-15 Classification: HYDROLASE Ligands: CDP, TS6, MG, PEG, GOL |
 |
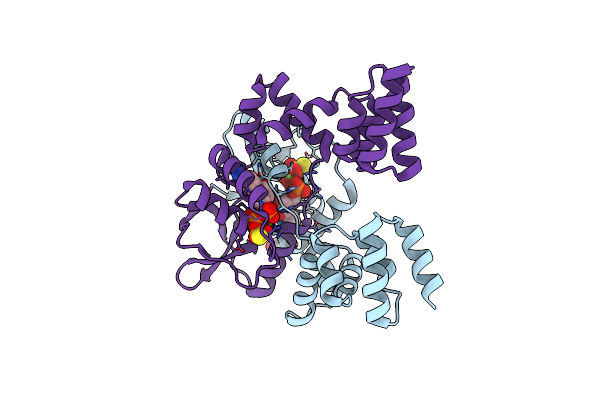
Crystal Structure Of Parb From Myxococcus Xanthus Bound To Cdp And Monothiophosphate
Organism: Myxococcus xanthus (strain dk 1622)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-08 Classification: DNA BINDING PROTEIN Ligands: GOL, PEG, TS6, CDP, MG |
 |
Crystal Structure Of A Parb Q52A Mutant From Myxococcus Xanthus Bound To Ctpys
Organism: Myxococcus xanthus (strain dk 1622)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-08 Classification: DNA BINDING PROTEIN Ligands: UFQ, GOL, MG |
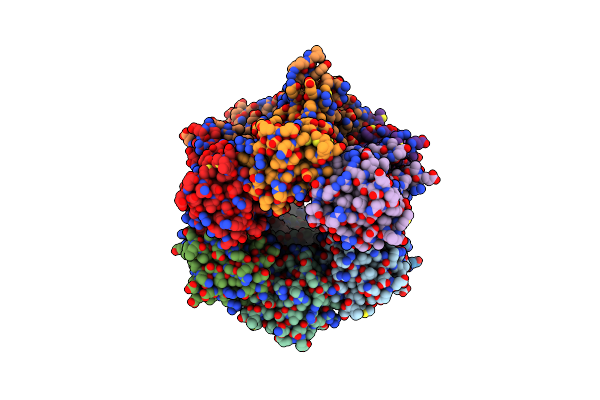 |
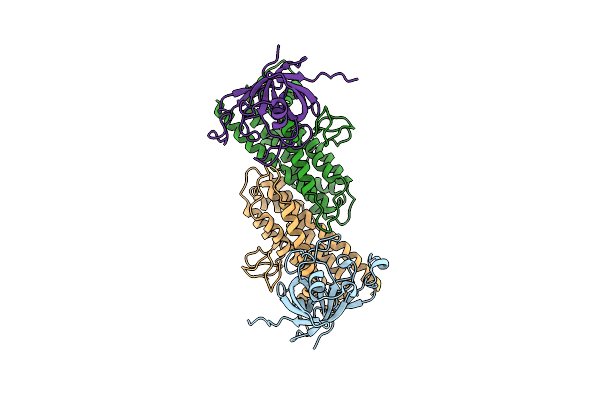
Organism: Halorhodospira halophila (strain dsm 244 / sl1)
Method: ELECTRON MICROSCOPY Release Date: 2021-07-07 Classification: HYDROLASE |
 |
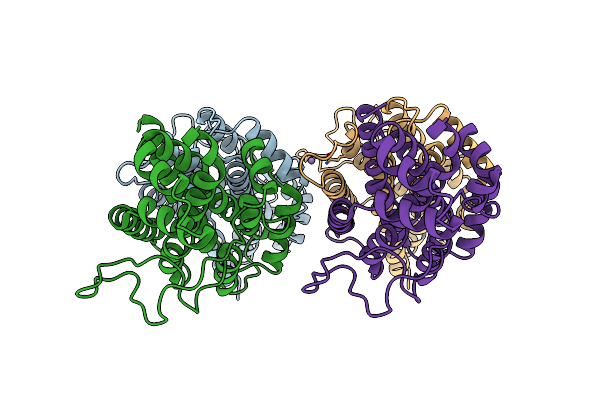
Structure Of The Ustilago Maydis Chorismate Mutase 1 In Complex With Kwl1-B From Zea Mays
Organism: Ustilago maydis 521, Zea mays
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-05-06 Classification: CELL INVASION |
 |
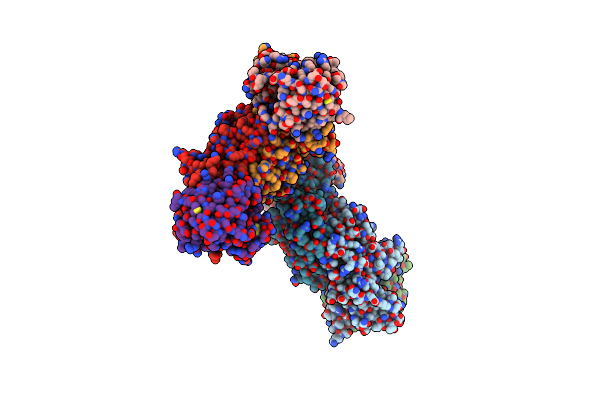
Organism: Ustilago maydis (strain 521 / fgsc 9021)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-01-16 Classification: CELL INVASION Ligands: MN |
 |
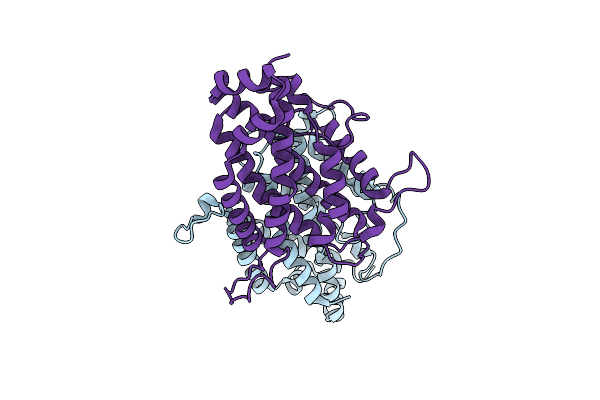
Structure Of The Ustilago Maydis Chorismate Mutase 1 In Complex With A Zea Mays Kiwellin
Organism: Ustilago maydis (strain 521 / fgsc 9021), Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-01-16 Classification: CELL INVASION Ligands: CIT |
 |
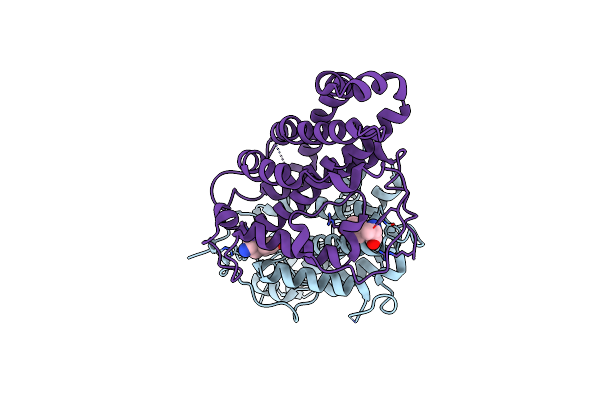
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-01-16 Classification: PLANT PROTEIN |
 |
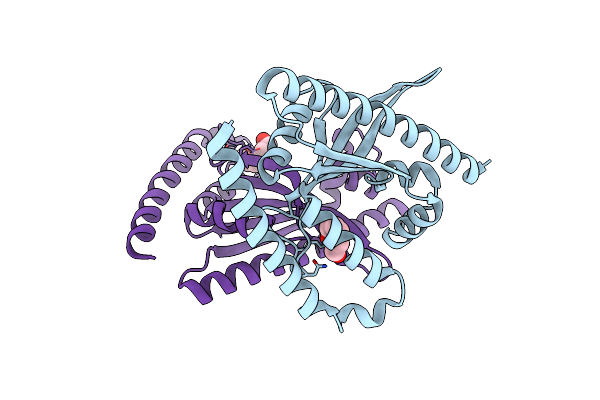
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-01-16 Classification: PLANT PROTEIN Ligands: TYR |
 |
Crystal Structure Of The Small Alarmone Synthethase 2 From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: PGE |
 |
Crystal Structure Of The Small Alarmone Synthethase 2 From Bacillus Subtilis
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-02-07 Classification: TRANSFERASE |
 |
Crystal Structure Of The Small Alarmone Synthethase 2 From Staphylococcus Aureus Bound To Ampcpp
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-02-07 Classification: TRANSFERASE Ligands: MG, APC |

