Search Count: 16
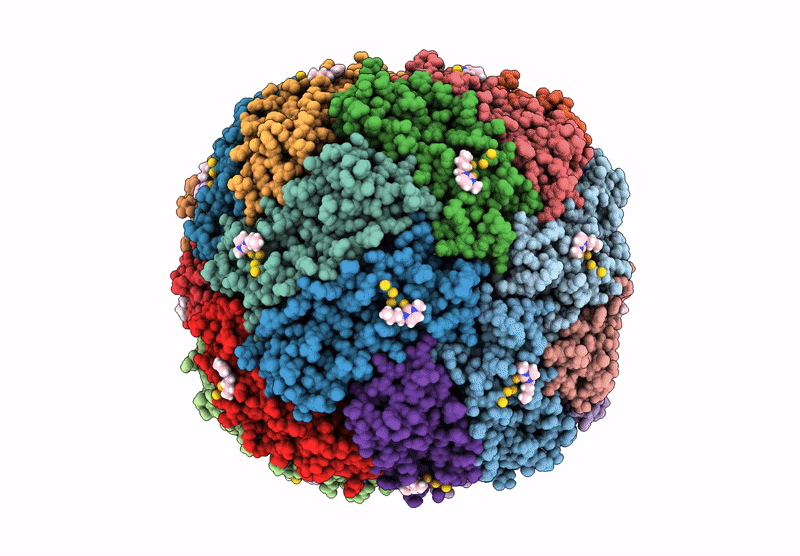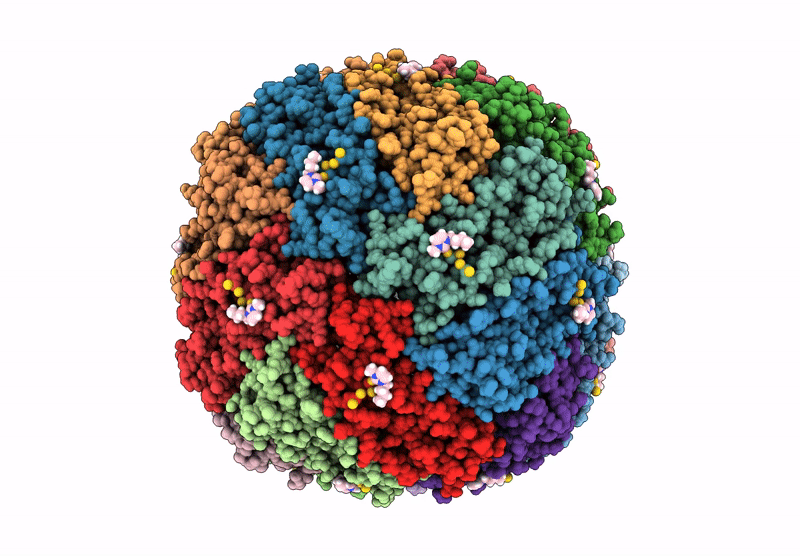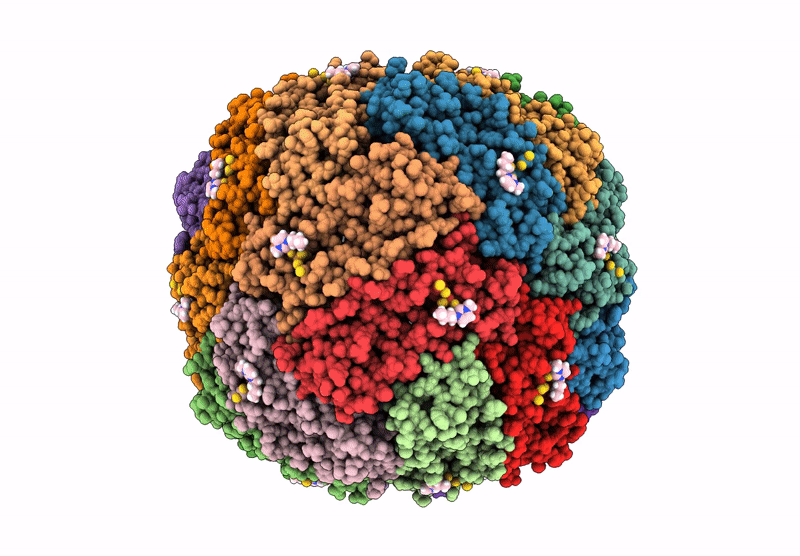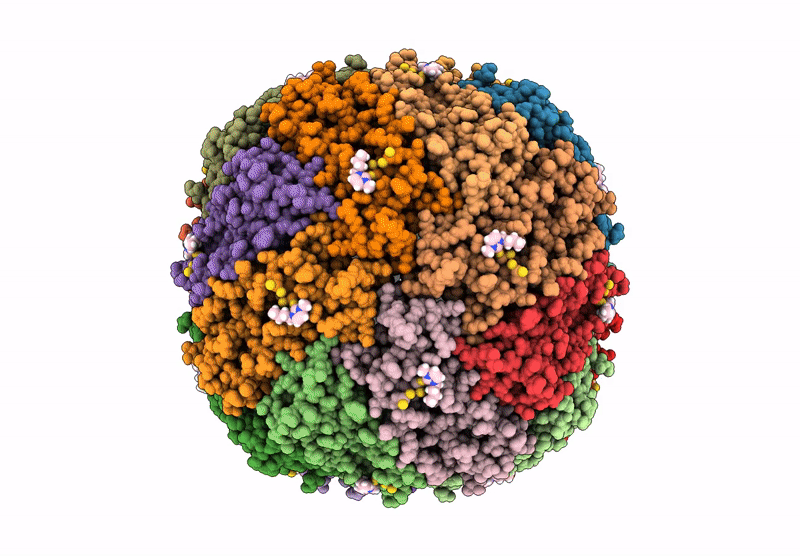 |
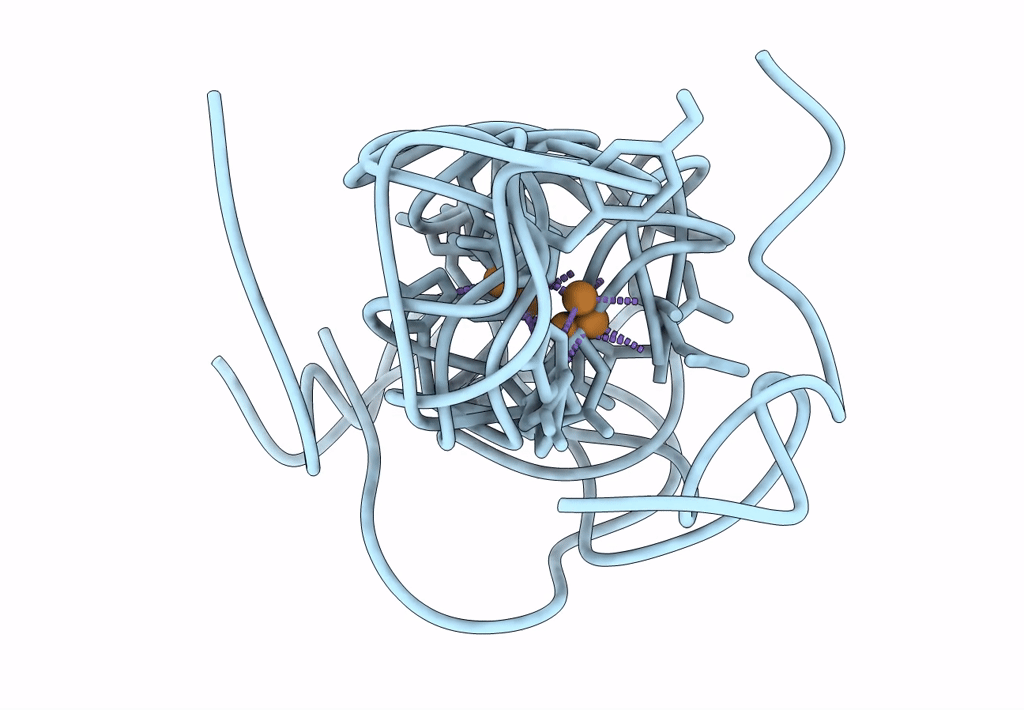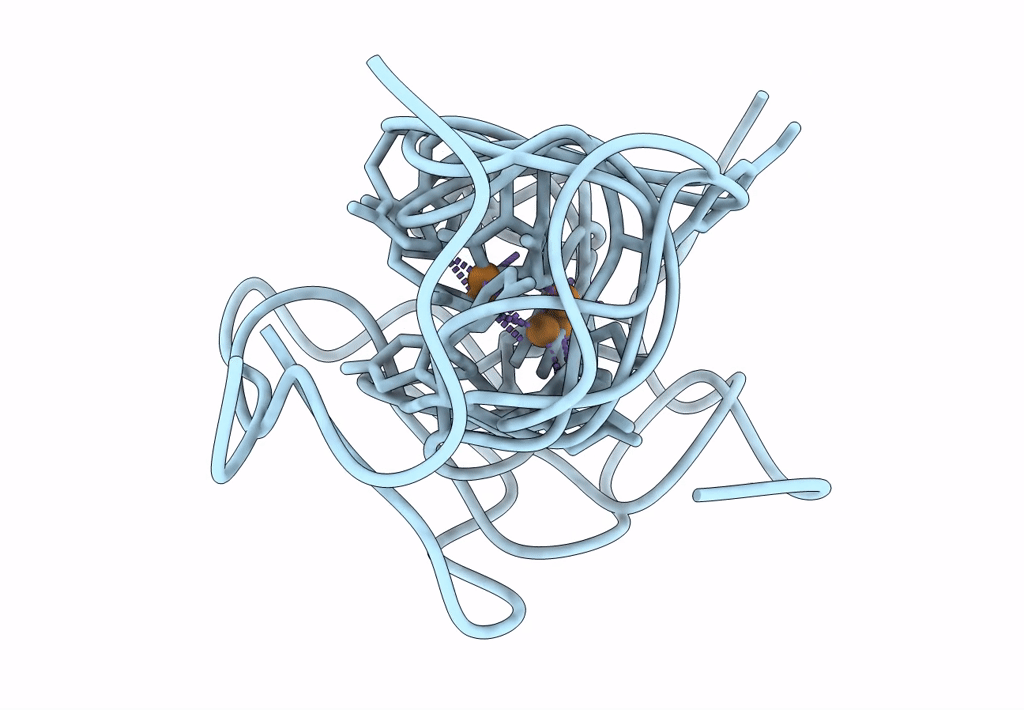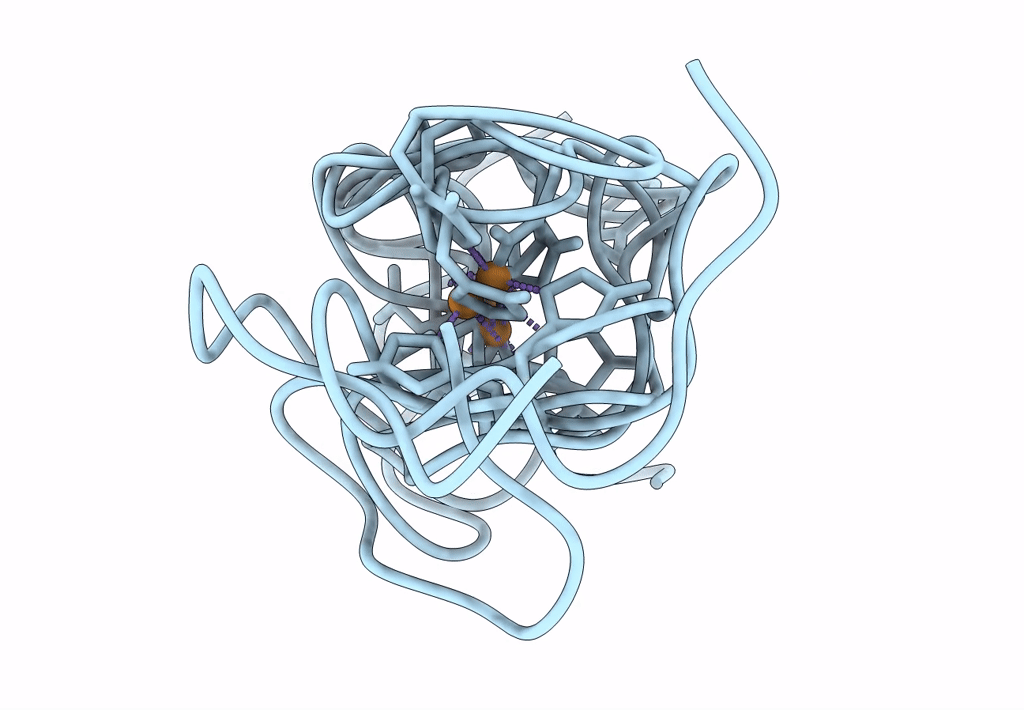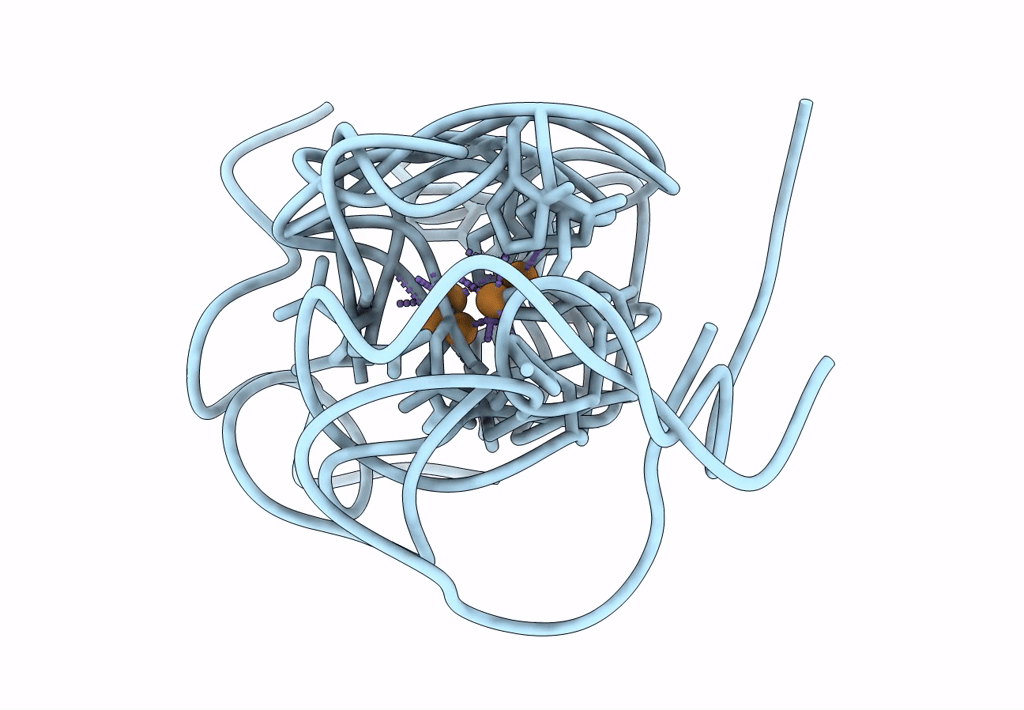
Structural Insights In The Huhf@Gold-Monocarbene Adduct: Aurophilicity Revealed In A Biological Context
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:1.51 Å Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: BM0, AU |
 |
Molecular Structure Of Cu(Ii)-Bound Amyloid-Beta Monomer Implicated In Inhibition Of Peptide Self-Assembly In Alzheimer'S Disease
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-08 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Molecular Structure Of Cu(Ii)-Bound Amyloid-Beta Monomer Implicated In Inhibition Of Peptide Self-Assembly In Alzheimer'S Disease
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2023-02-01 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
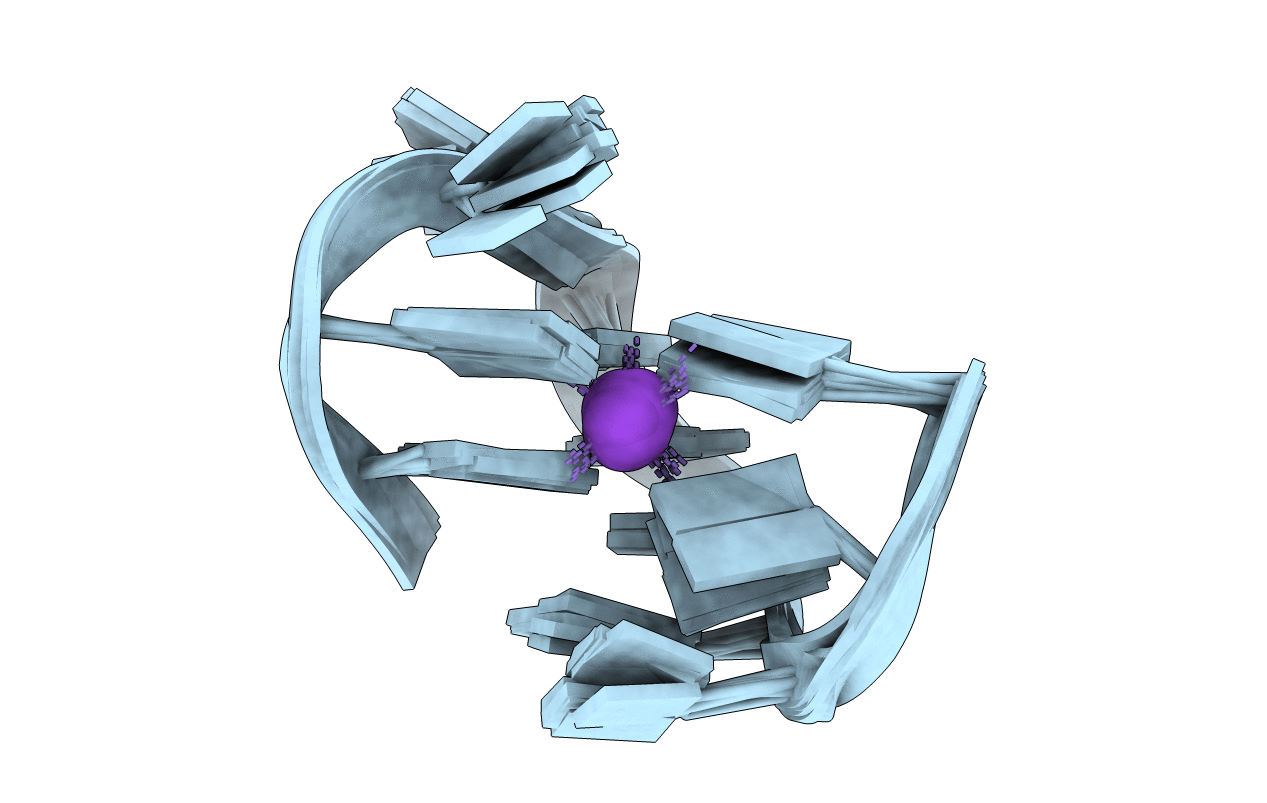 |
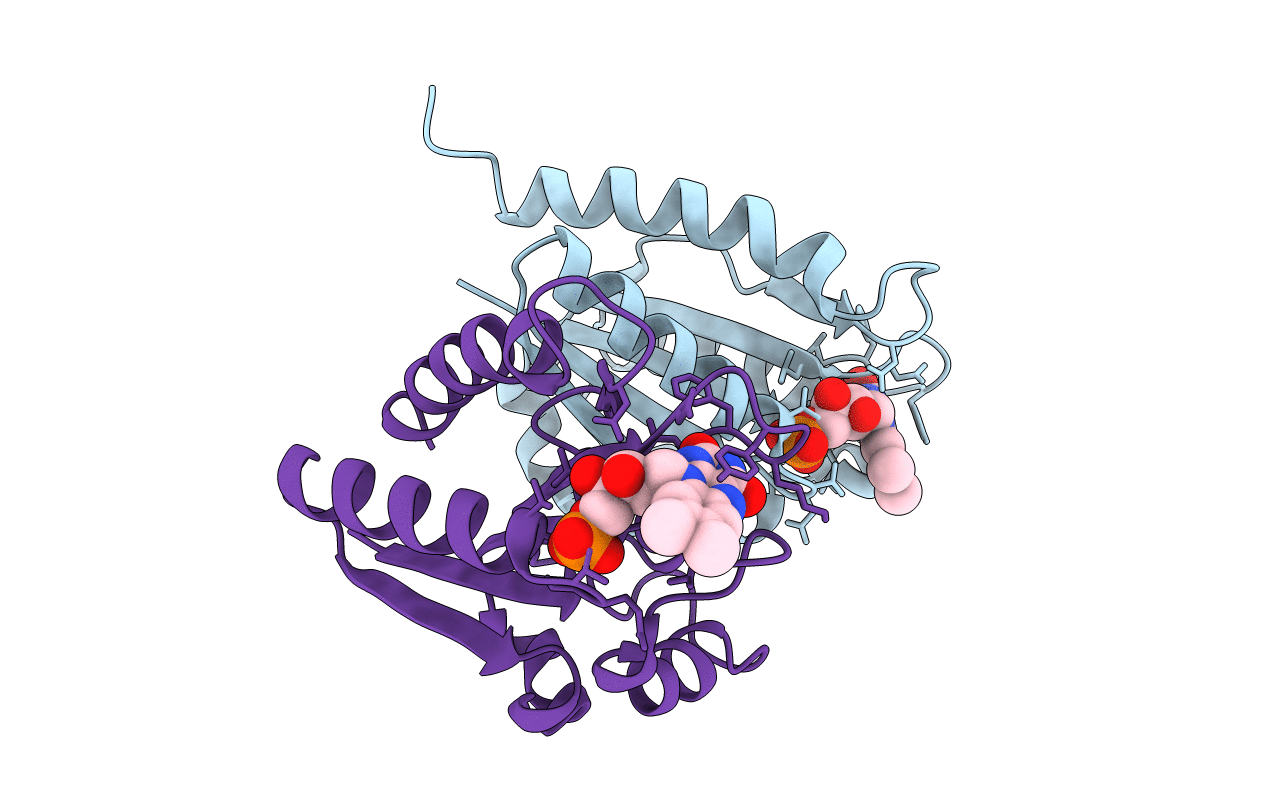 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-04-17 Classification: OXIDOREDUCTASE Ligands: FMN |
 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor Paramethoxy-Sulfonyl-Glycine Hydroxamate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-10-06 Classification: HYDROLASE Ligands: ZN, CA, Z79 |
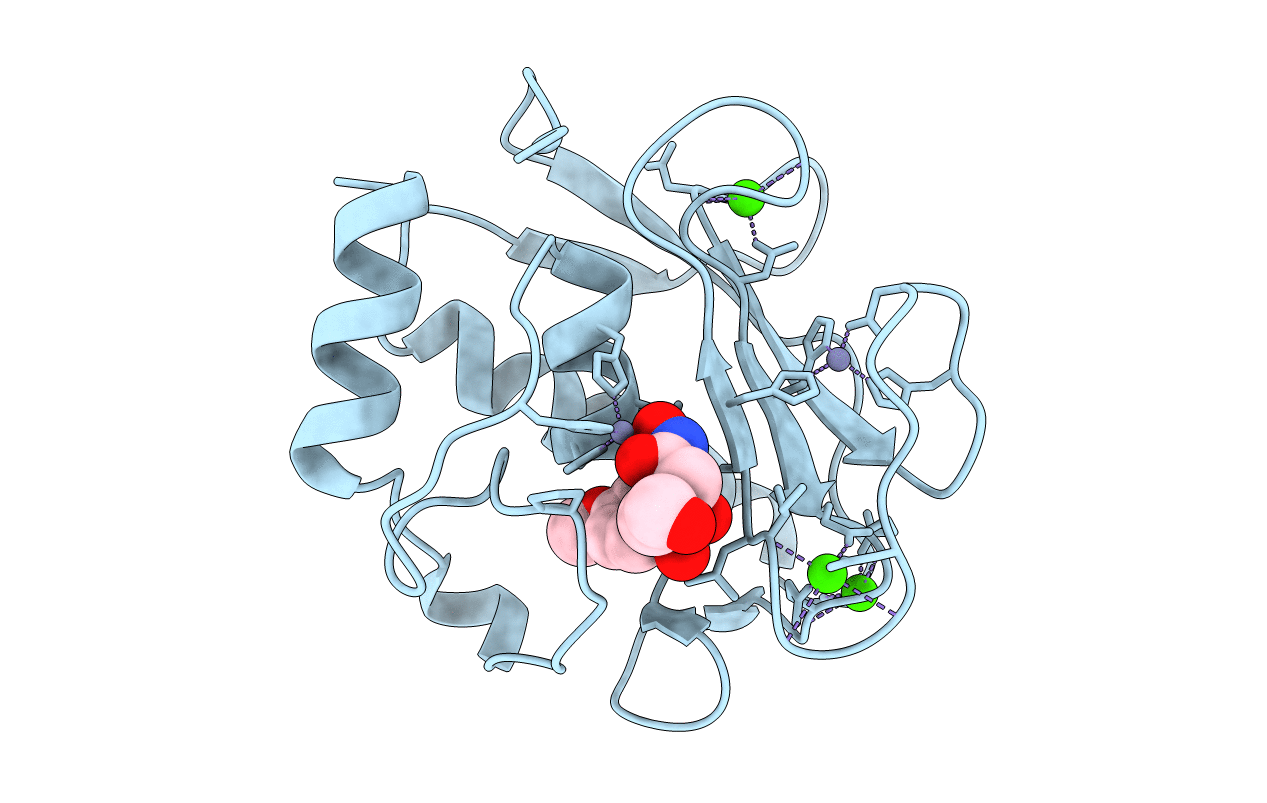 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor N-Hydroxy-2-(N-(2-Hydroxyethyl)4-Methoxyphenylsulfonamido)Acetamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-07-28 Classification: HYDROLASE Ligands: ZN, CA, NHK |
 |
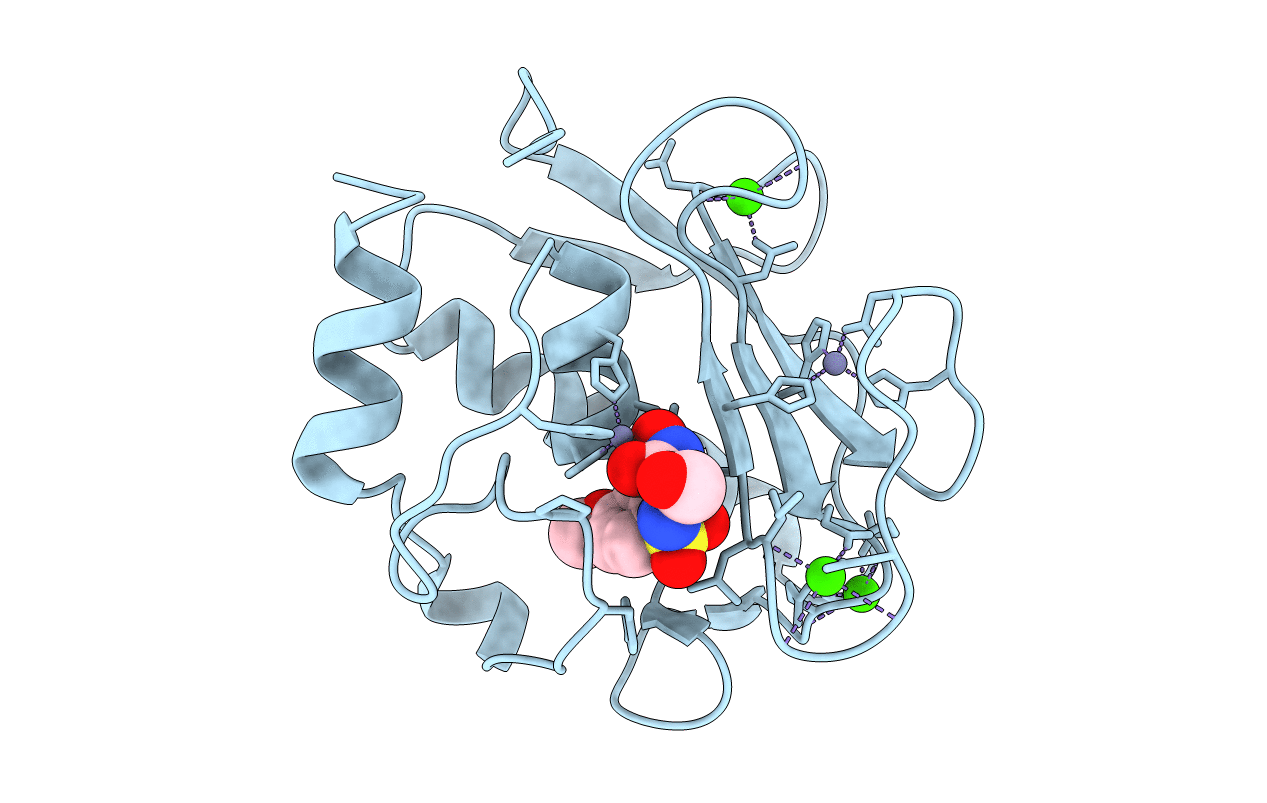
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor (S)-N-(2,3-Dihydroxypropyl)-4-Methoxy-N-(2-Nitroso-2-Oxoethyl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS1 |
 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor (R)-N-(3-Hydroxy-1-Nitroso-1-Oxopropan-2-Yl)-4-Methoxybenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS3 |
 |
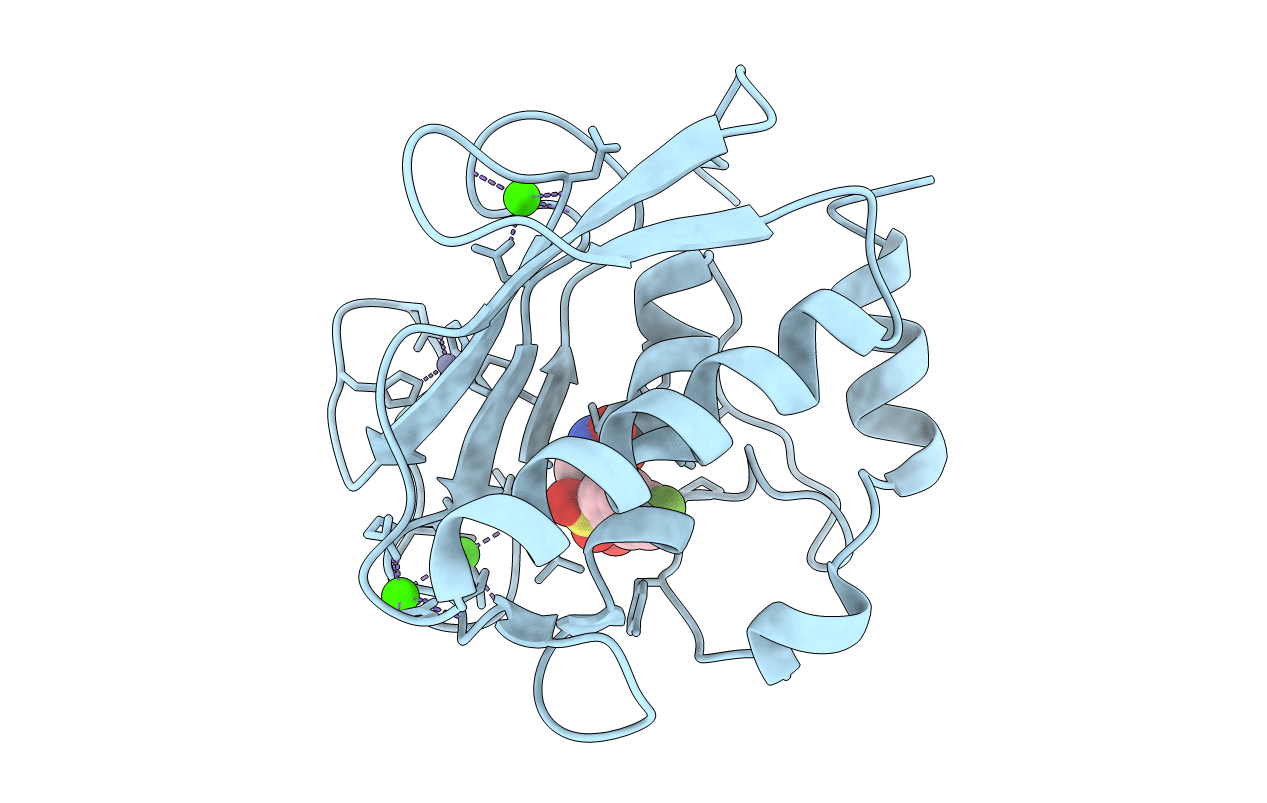
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor N-(2-Nitroso-2-Oxoethyl)Biphenyl-4-Sulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS4 |
 |
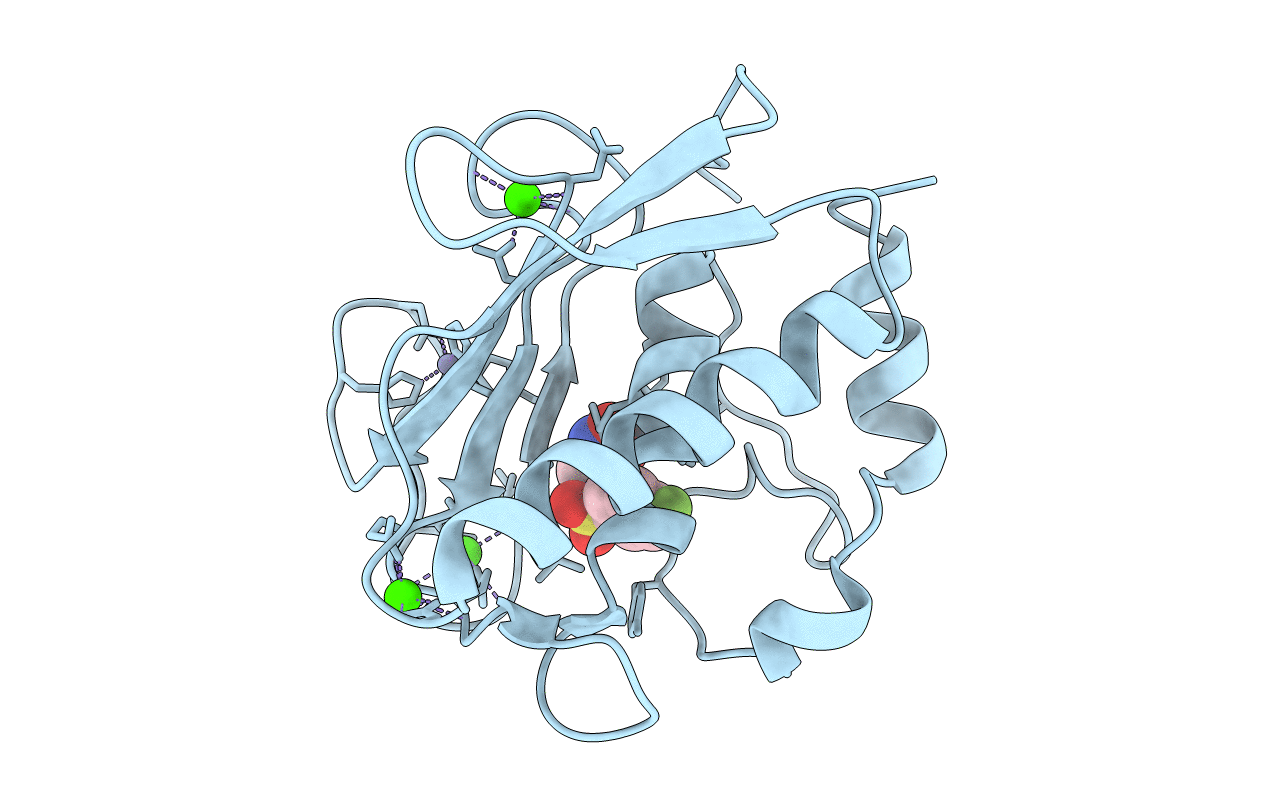
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor 4-Fluoro-N-(2-Hydroxyethyl)-N-(2-Nitroso-2-Oxoethyl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS5 |
 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor 4-Fluoro-N-(2-Nitroso-2-Oxoethyl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS6 |
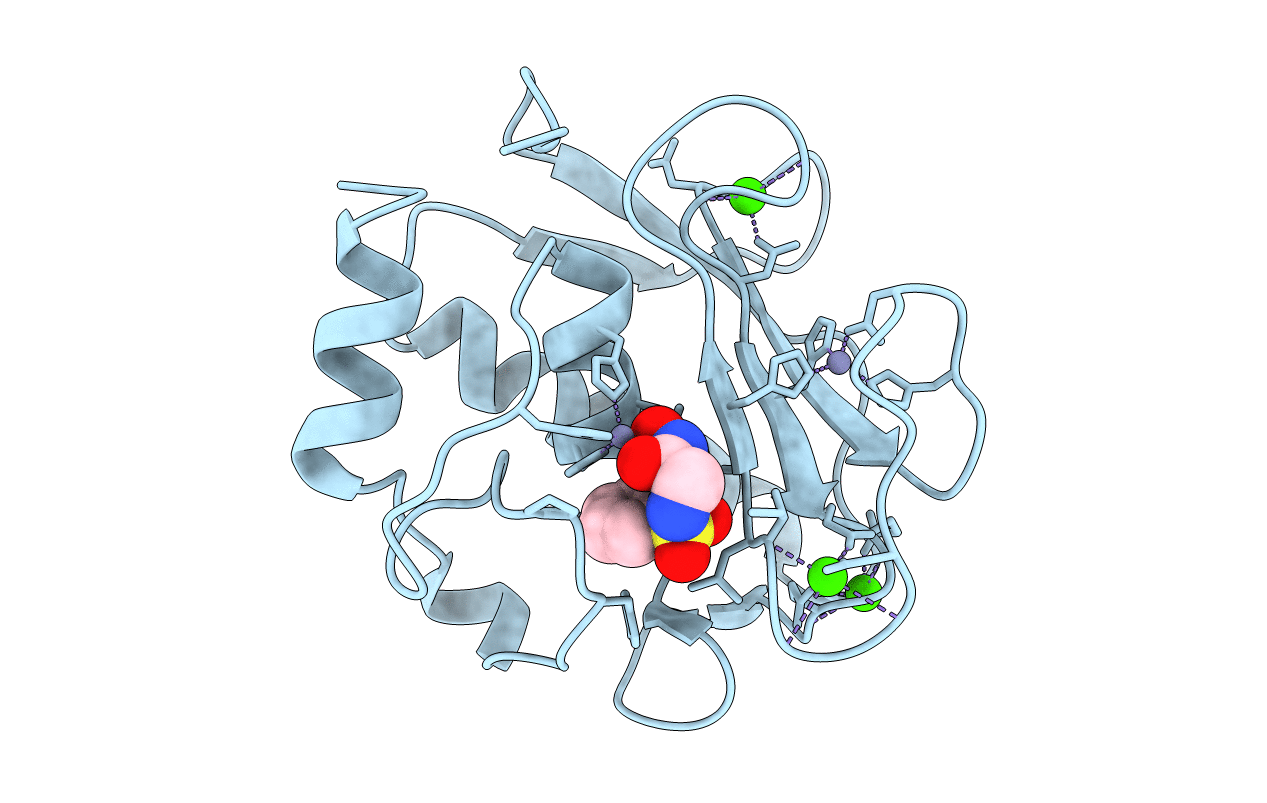 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor N-(2-Nitroso-2-Oxoethyl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: ZN, CA, HS7 |
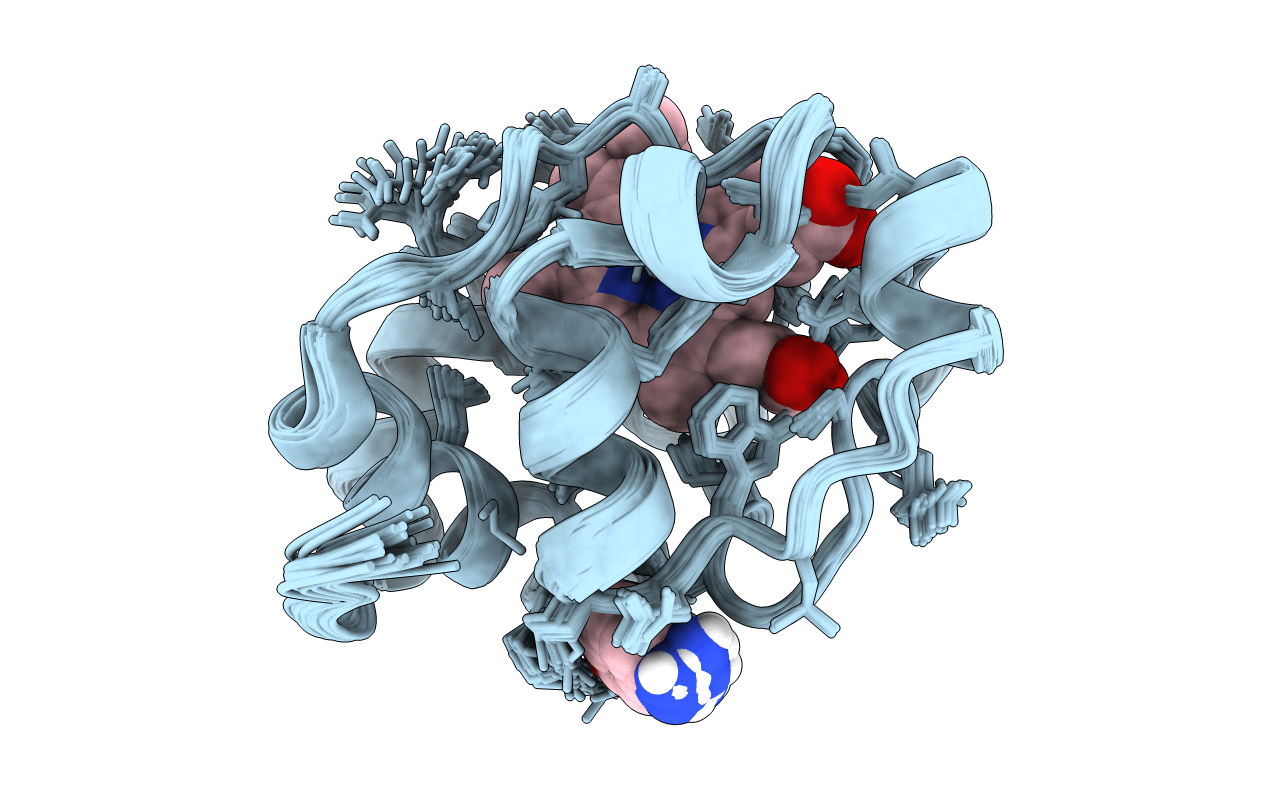 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2007-04-24 Classification: ELECTRON TRANSPORT Ligands: HEC, 4NL |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2006-09-26 Classification: ELECTRON TRANSPORT Ligands: HEC |

