Search Count: 30
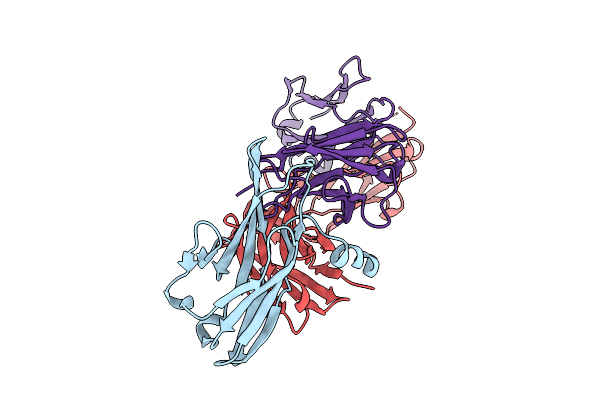 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
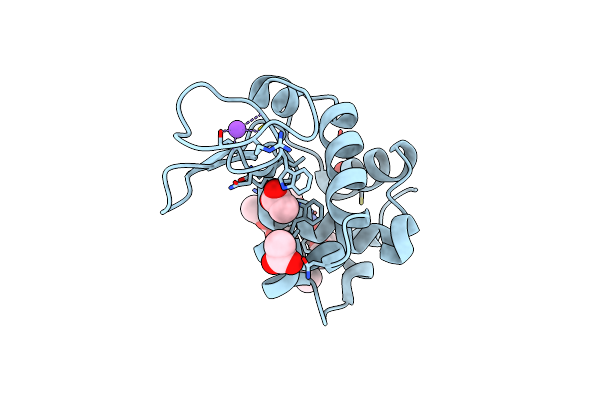 |
X-Ray Structure Of Lysozyme Obtained Upon Reaction With The Dioxidovanadium(V) Complex With Furan-2-Carboxylic Acid (3-Ethoxy-2-Hydroxybenzylidene)Hydrazide
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2024-12-18 Classification: HYDROLASE Ligands: CL, NA, ACT, VVB, A1IOM |
 |
X-Ray Structure Of Lysozyme Obtained Upon Reaction With The Dioxidovanadium(V) Complex With (E)-N'-(1-(2-Hydroxy-5-Methoxyphenyl)Ethylidene)Furan-2-Carbohydrazide)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2024-12-18 Classification: HYDROLASE |
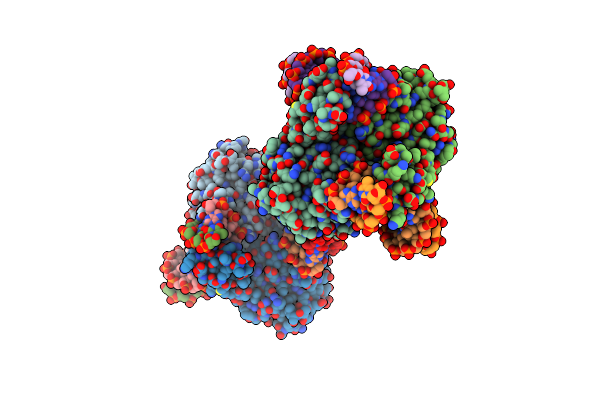 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrates
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: EPE, SFG |
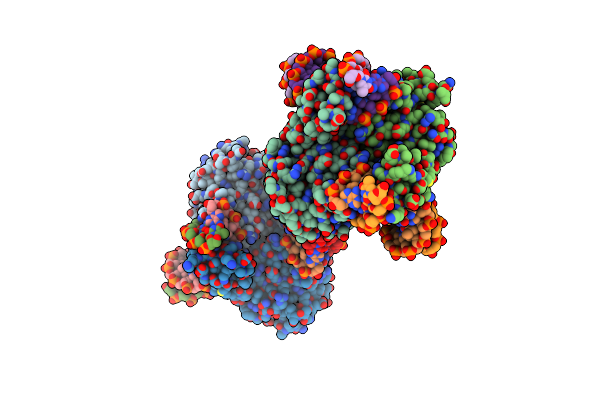 |
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtatac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
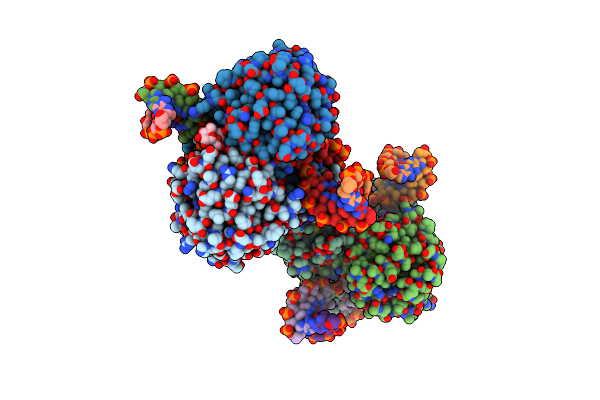
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtaaac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
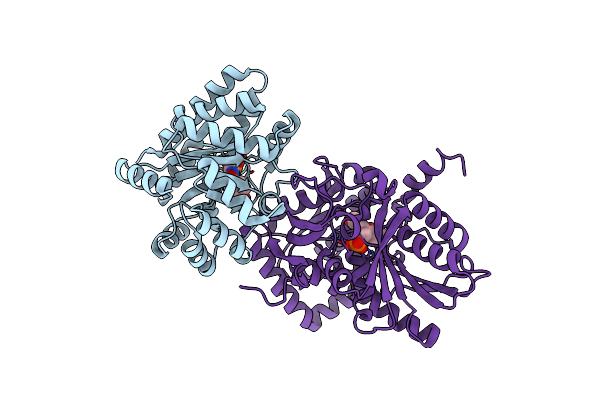
Crystal Structure Of Dna N6-Adenine Methyltransferase M.Bcejiv From Burkholderia Cenocepacia In Complex With Duplex Dna Substrate Containing Gtttac As Recognition Sequence
Organism: Burkholderia cenocepacia
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2024-09-11 Classification: DNA BINDING PROTEIN Ligands: SFG |
 |
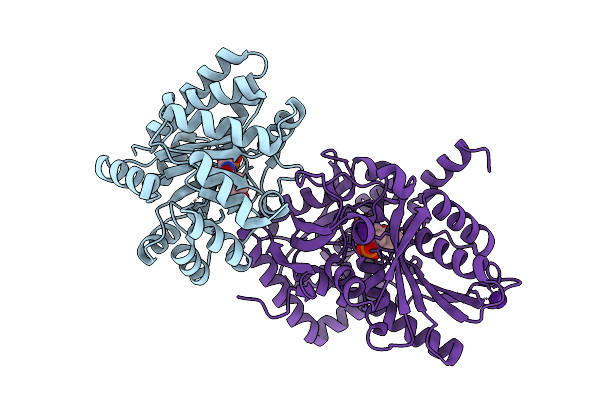
Crystal Structure Of Tryptophan Synthase In Complex With F9, Nh4+, Ph7.8 - Alpha Aminoacrylate Form - E(A-A)
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-12-29 Classification: LYASE/INHIBITOR Ligands: F9F, 0JO, NH4 |
 |
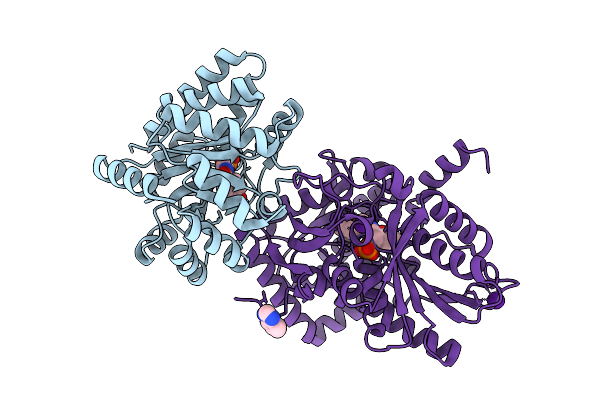
Crystal Structure Of Tryptophan Synthase In Complex With F9, Cs+, Ph7.8 - Alpha Aminoacrylate Form - E(A-A)
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-12-29 Classification: LYASE/INHIBITOR Ligands: F9F, 0JO, CS |
 |
Crystal Structure Of Tryptophan Synthase In Complex With F9, Cs+, Benzimidazole, Ph7.8 - Alpha Aminoacrylate Form - E(A-A)(Bzi)
Organism: Salmonella typhimurium, Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-29 Classification: LYASE/INHIBITOR Ligands: F9F, 0JO, CS, BZI |
 |
Atp-Competitive Partial Antagonists-'Pair'S-Rheostatically Modulate Ire1Alpha'S Kinase Helix-Alphac To Segregate Its Rnase-Mediated Biological Outputs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-29 Classification: UNKNOWN FUNCTION Ligands: U4N, GOL, EDO, PEG, PGE |
 |
Organism: Gallus gallus, Chara corallina
Method: ELECTRON MICROSCOPY Release Date: 2021-01-13 Classification: CONTRACTILE PROTEIN Ligands: ADP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:9.80 Å Release Date: 2019-06-19 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:10.70 Å Release Date: 2019-01-23 Classification: TRANSCRIPTION |
 |
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-03-09 Classification: LIGASE Ligands: ATP, SO4, MG |
 |
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2016-03-09 Classification: LIGASE Ligands: SO4, MG |
 |
A Bimolecular Anti-Parallel-Stranded Oxytricha Nova Telomeric Quadruplex In Complex With A 3,6-Disubstituted Acridine Bsu-6042 In A Large Unit Cell
Method: X-RAY DIFFRACTION
Resolution:2.20 Å Release Date: 2008-12-02 Classification: DNA Ligands: K, NC5, SPM, NCI |
 |
A Bimolecular Anti-Parallel-Stranded Oxytricha Nova Telomeric Quadruplex In Complex With A 3,6-Disubstituted Acridine Bsu-6066
Method: X-RAY DIFFRACTION
Resolution:1.78 Å Release Date: 2008-10-21 Classification: DNA Ligands: K, NCQ |
 |
A Bimolecular Anti-Parallel-Stranded Oxytricha Nova Telomeric Quadruplex In Complex With A 3,6-Disubstituted Acridine Bsu-6038
Method: X-RAY DIFFRACTION
Resolution:2.30 Å Release Date: 2008-10-14 Classification: DNA Ligands: K, NCE |
 |
A Bimolecular Anti-Parallel-Stranded Oxytricha Nova Telomeric Quadruplex In Complex With A 3,6-Disubstituted Acridine Bsu-6042 In Small Unit Cell
Method: X-RAY DIFFRACTION
Resolution:2.20 Å Release Date: 2008-10-14 Classification: DNA Ligands: K, NCI |

