Search Count: 33
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, O |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, PEG, SO4, CL |
 |
Organism: Vibrio cholerae o395
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-04-17 Classification: SUGAR BINDING PROTEIN Ligands: G6P |
 |
Organism: Vibrio cholerae o395
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-02 Classification: TRANSCRIPTION Ligands: EDO, SO4 |
 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
 |
Crystal Structure Of The N-Terminal Domain Of Mutants Of Human Apolipoprotein-E (Apoe)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: LIPID TRANSPORT |
 |
Crystal Structure Of Vpsr Display Novel Dimeric Architecture And C-Di-Gmp Binding: Mechanistic Implications In Oligomerization, Atpase Activity And Dna Binding.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-04-06 Classification: TRANSCRIPTION Ligands: GTP |
 |
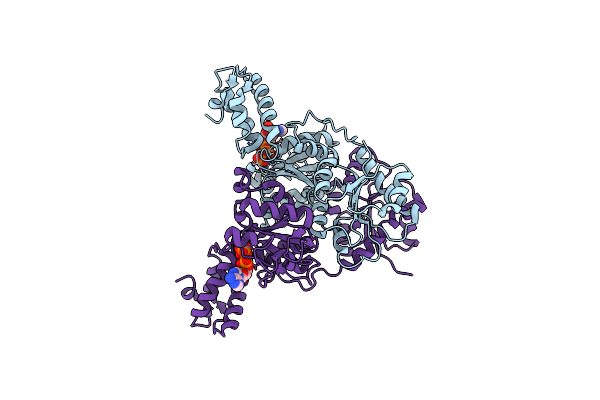
Crystal Structure Of Vpsr Display Novel Dimeric Architecture And C-Di-Gmp Binding: Mechanistic Implications In Oligomerization, Atpase Activity And Dna Binding.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2022-04-06 Classification: TRANSCRIPTION Ligands: SO4 |
 |
Crystal Structure Of Vpsr Display Novel Dimeric Architecture And C-Di-Gmp Binding: Mechanistic Implications In Oligomerization, Atpase Activity And Dna Binding.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2022-04-06 Classification: TRANSCRIPTION Ligands: ATP |
 |
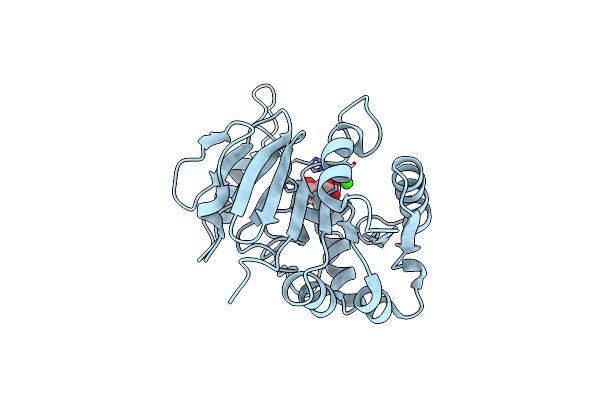
Crystal Structure Of Vpsr Display Novel Dimeric Architecture And C-Di-Gmp Binding: Mechanistic Implications In Oligomerization, Atpase Activity And Dna Binding.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2022-04-06 Classification: TRANSCRIPTION Ligands: SO4, C2E |
 |
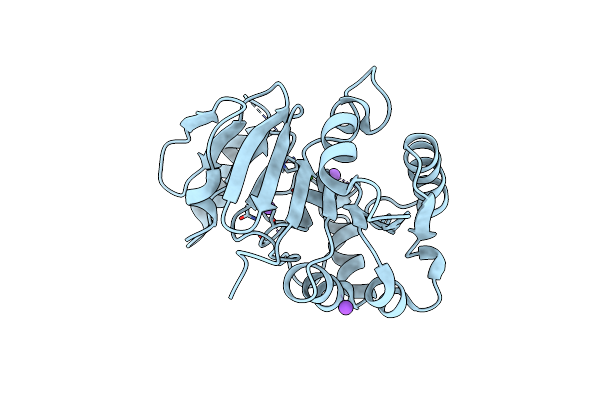
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: ASP, CA |
 |
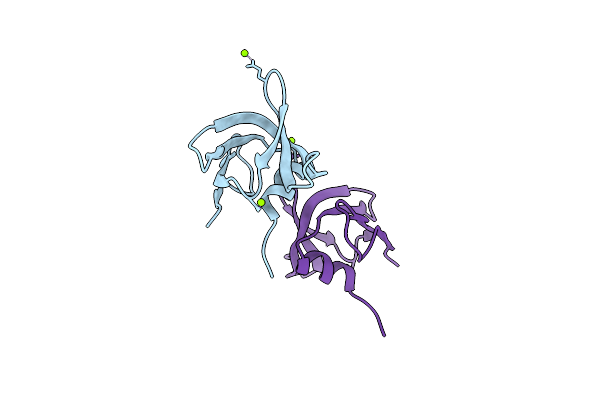
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-06-09 Classification: HYDROLASE Ligands: CA, NA |
 |
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-12-11 Classification: TRANSCRIPTION Ligands: MG |
 |
Crystal Structure Of Pyrrolidone Carboxylate Peptidase I From Deionococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-01-16 Classification: HYDROLASE |
 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: LIPID BINDING PROTEIN |
 |
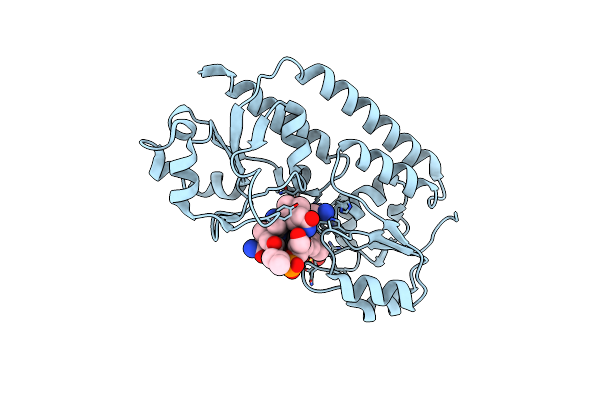
Crystal Structure Of The S37A Mutant Of Apo-Acyl Carrier Protein From Leishmania Major
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: LIPID BINDING PROTEIN |
 |
Crystal Structure Of Periplasmic Vitamin B12 Binding Protein Btuf Of Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2018-09-19 Classification: TRANSPORT PROTEIN Ligands: CNC, SO4 |
 |
Crystal Structure Of Low Molecular Weight Phosphotyrosine Phosphatase (Vclmwptp-2) From Vibrio Choleraeo395
Organism: Vibrio cholerae serotype o1 (strain atcc 39541 / classical ogawa 395 / o395)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-09-19 Classification: HYDROLASE Ligands: MPO, SO4, GOL |
 |
N-Terminal Domain Of Fact Complex Subunit Spt16 From Eremothecium Gossypii (Ashbya Gossypii)
Organism: Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-15 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of A Novel Xaa-Pro Dipeptidase From Deinococcus Radiodurans
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-10-18 Classification: HYDROLASE Ligands: ZN, PO4, ACT, CL, EDO |

