Search Count: 13
 |
Organism: Oryza sativa subsp. indica
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: FAD, NRF, EPE, IMD, TRS |
 |
Organism: Pantoea ananatis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2012-10-10 Classification: OXIDOREDUCTASE Ligands: EDO, CL |
 |
Organism: Brevibacterium sterolicum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-12-11 Classification: OXIDOREDUCTASE Ligands: MN, CAC, FAD, GOL |
 |
Crystal Structure Of D-Amino Acid Oxidase In Complex With Two Anthranylate Molecules
Organism: Rhodosporidium toruloides
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-02-27 Classification: OXIDOREDUCTASE Ligands: FAD, BE2 |
 |
Organism: Brevibacterium sterolicum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-08-08 Classification: OXIDOREDUCTASE Ligands: EDO, FAD, MN, CAC |
 |
Crystal Structure Analysis Of D-Amino Acid Oxidase In Complex With L-Lactate
Organism: Rhodosporidium toruloides
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2000-11-22 Classification: OXIDOREDUCTASE Ligands: FAD, LAC |
 |
D-Amino Acid Oxidase: Structure Of Substrate Complexes At Very High Resolution Reveal The Chemical Reacttion Mechanism Of Flavin Dehydrogenation
Organism: Rhodosporidium toruloides
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2000-11-22 Classification: OXIDOREDUCTASE Ligands: FLA, FAD |
 |
D-Amino Acic Oxidase In Complex With D-Alanine And A Partially Occupied Biatomic Species
Organism: Rhodosporidium toruloides
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2000-11-22 Classification: OXIDOREDUCTASE Ligands: FAD, DAL, PER, GOL |
 |
Crystal Structure Of L-Amino Acid Oxidase From Calloselasma Rhodostoma Complexed With Citrate
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-24 Classification: OXIDOREDUCTASE Ligands: NAG, CIT, FAD |
 |
Crystal Structure Of L-Amino Acid Oxidase From Calloselasma Rhodostoma, Complexed With Three Molecules Of O-Aminobenzoate.
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-24 Classification: OXIDOREDUCTASE Ligands: NAG, BE2, FAD |
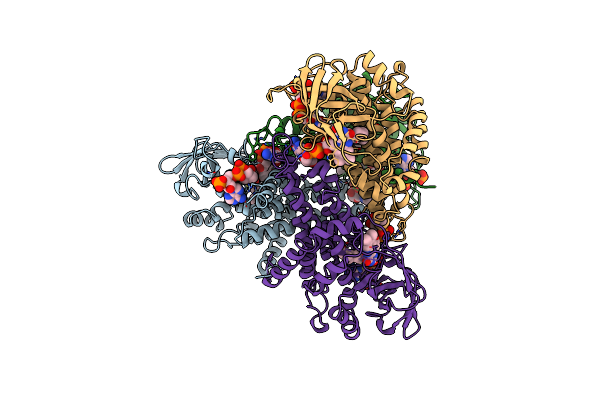 |
Structure Of T255E, E376G Mutant Of Human Medium Chain Acyl-Coa Dehydrogenase Complexed With Octanoyl-Coa
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1997-11-12 Classification: ELECTRON TRANSFER Ligands: CO8, FAD |
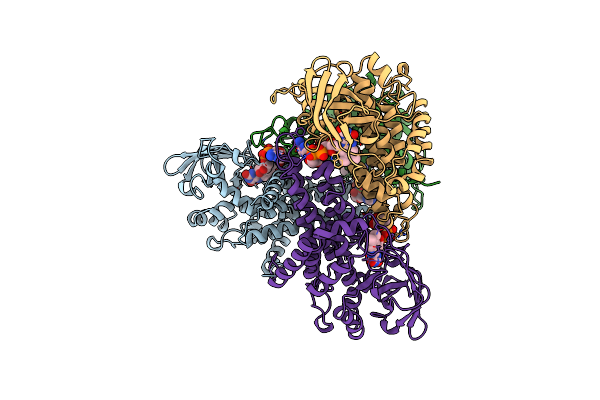 |
Structure Of T255E, E376G Mutant Of Human Medium Chain Acyl-Coa Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1997-06-16 Classification: ELECTRON TRANSFER Ligands: FAD |
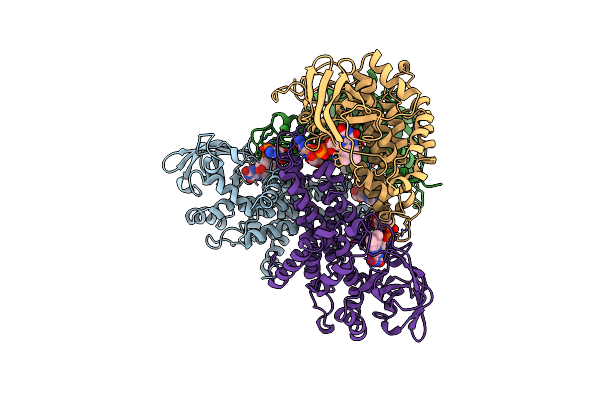 |
Structure Of T255E, E376G Mutant Of Human Medium Chain Acyl-Coa Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 1997-06-16 Classification: ELECTRON TRANSFER Ligands: FAD |

