Search Count: 80
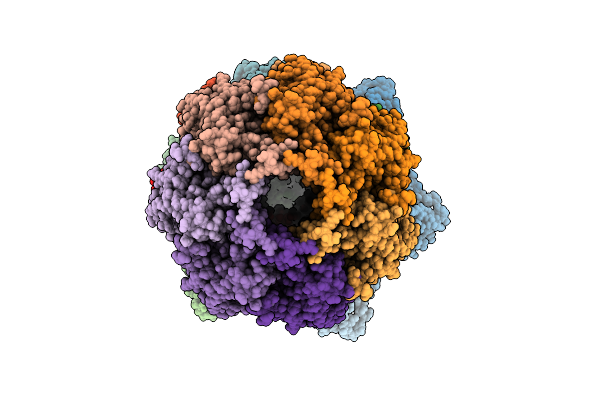 |
Cryo-Em Structure Of The Clpxp Aaa+ Protease Bound To Lambdao-Tagged Arc In A Recognition Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CHAPERONE Ligands: AGS, MG |
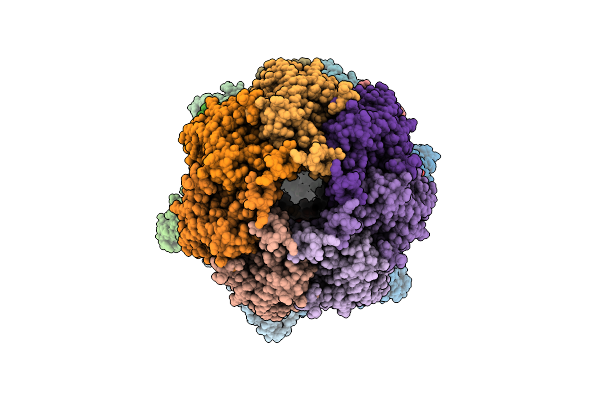 |
Cryo-Em Structure Of The Clpxp Aaa+ Protease Bound To An Unidentified Portion Of Lambdao-Tagged Arc Substrate Within A Translocation Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CHAPERONE Ligands: AGS, MG |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: RETINOL BINDING PROTEIN Ligands: A1CKW, ACT |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-9
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: RETINOL BINDING PROTEIN Ligands: ACT, A1CK1 |
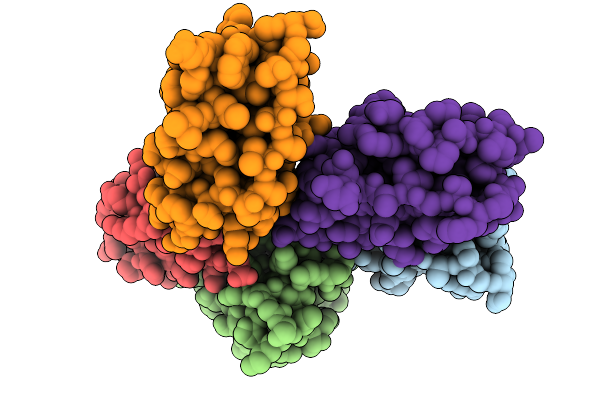 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIPID BINDING PROTEIN |
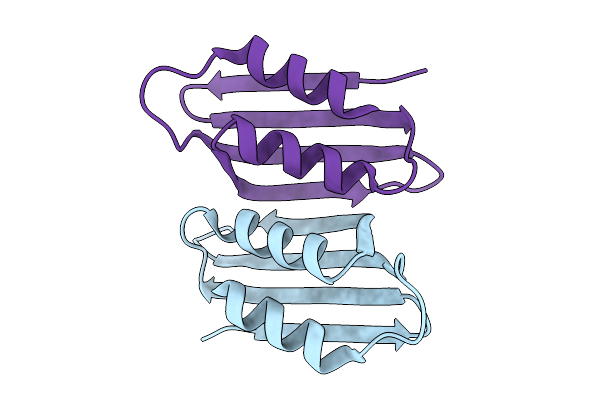 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: LIPID BINDING PROTEIN |
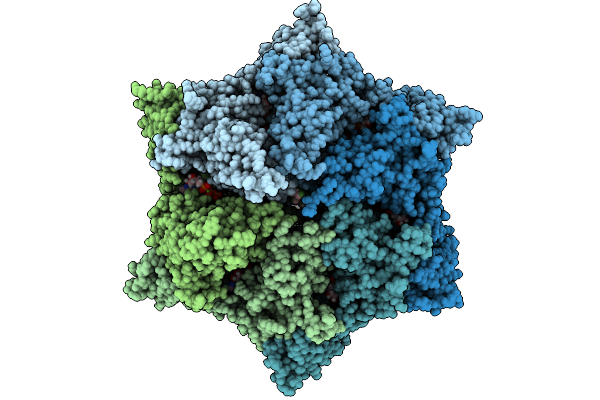 |
Cryo-Em Structure Of The Endogenous Clpp1/Clpp2 Heterocomplex From Pseudomonas Aeruginosa Bound To The Aaa+ Clpx Unfoldase.
Organism: Pseudomonas aeruginosa, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: CHAPERONE Ligands: MG, ATP, ADP |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-7
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CKU, ACT |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CKV, ACT |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CKX, ACT |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CKY, ACT, GOL |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CKZ, ACT |
 |
Q108K:K40L:T51V:T53S:R58W:Y19W:L117E Mutant Of Hcrbpii Bound To Fluorophore Td-1V-10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: RETINOL BINDING PROTEIN Ligands: A1CK0, ACT |
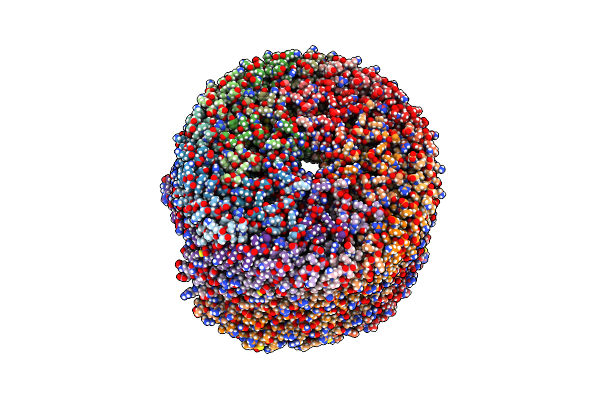 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CHAPERONE |
 |
Cryo-Em Structure Of A Nautilus-Like Hflk/C Assembly In Complex With Ftsh Aaa Protease
Organism: Escherichia coli bl21
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: CHAPERONE |
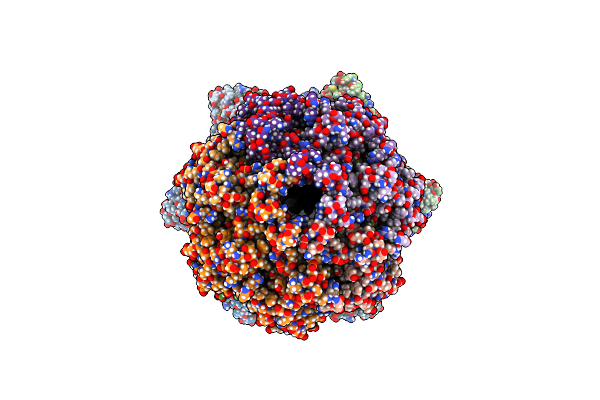 |
Cryo-Em Structure Of A Proteolytic Clpxp Aaa+ Machine Poised To Unfold A Branched-Degron Dhfr-Ssra Substrate Bound With Mtx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CHAPERONE Ligands: ATP, MG, ADP, MTX |
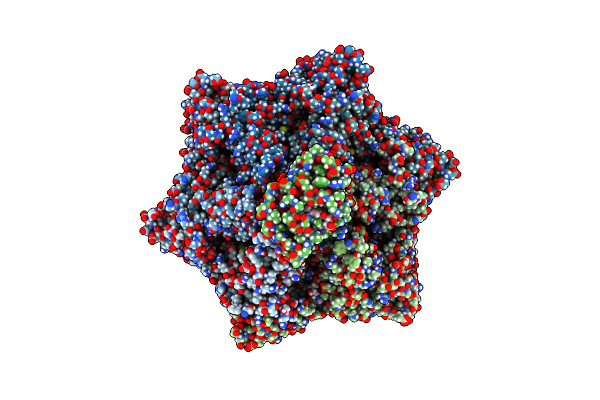 |
Cryo-Em Structure Of A Proteolytic Clpxp Aaa+ Machine Poised To Unfold A Linear-Degron Dhfr-Ssra Substrate Bound With Mtx
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CHAPERONE Ligands: ATP, MG, ADP, MTX |
 |
Cryo-Em Structure Of A Proteolytic Clpxp Aaa+ Machine Translocating A Portion Of A Branched-Degron Dhfr Substrate
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: CHAPERONE Ligands: ATP, MG, ADP |
 |
Q108K:K40L:T53A:R58F Mutant Of Hcrbpii Bound To Synthetic Fluorophore Td-1V
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-04-03 Classification: RETINOL BINDING PROTEIN Ligands: ACT, A1AEQ |
 |
Q108K:K40L:T51V:T53S:R58Y Mutant Of Hcrbpii Bound To Synthetic Fluorophore Td-1V
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2024-04-03 Classification: RETINOL BINDING PROTEIN Ligands: A1AEQ, ACT |

