Search Count: 150
 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V90P(Nuoe)
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, NA, SO4, FMN, SF4 |
 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V136M(Nuoe)
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, CL, SO4, SF4, FMN, GOL, NA |
 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Mutation V136M(Nuoe), Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, CL, SO4, NA, SF4, FMN, NAD, GOL |
 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Double Mutation V90P And V136M(Nuoe), Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, SF4, FMN, NAD, NA, BU3, CL |
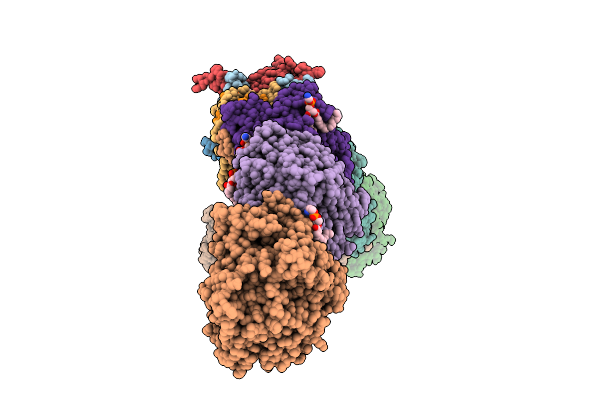 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: 3PE, SF4, FES, FMN, CA, CL |
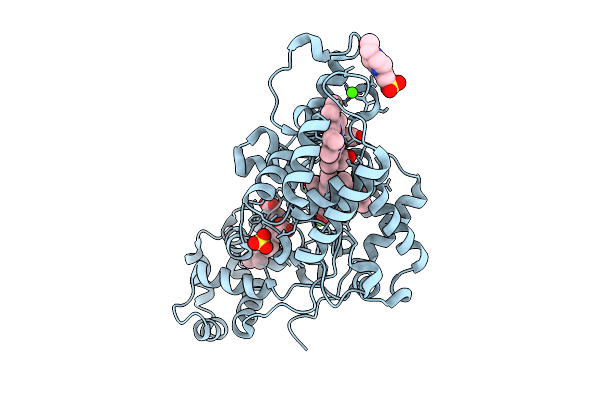 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: HEC, HEM, CA, CL, EPE, SO4 |
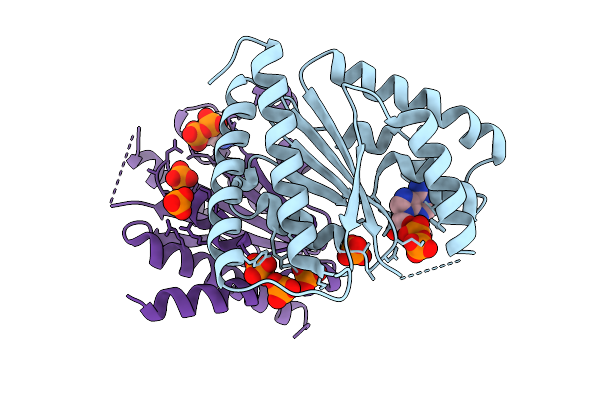 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Adp (Form I)
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: ADP, PO4 |
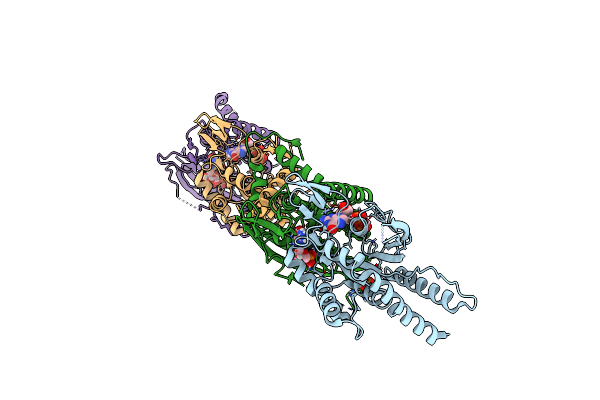 |
Crystal Structure Of Septin Complex Shs1-Cdc12-Cdc3-Cdc10 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-02-05 Classification: STRUCTURAL PROTEIN Ligands: GDP, MG, SO4 |
 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis In Apo Form
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: PO4, SIN |
 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Amp
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: CL, AMP, PO4 |
 |
Crystal Structure Of Polyphosphate Kinase 2-Ii (Ppk2-Ii) From Lysinibacillus Fusiformis Bound To Tmp
Organism: Lysinibacillus fusiformis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-05 Classification: TRANSFERASE Ligands: TMP, PO4 |
 |
Crystal Structure Of Sgvm Methyltransferase In Complex With Alpha-Ketoleucine And Zn2+ Ion
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: COI, CL, MG, ZN |
 |
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-07-24 Classification: TRANSFERASE Ligands: ZN, COI, CL, SAM |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Native-Form
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Ketoarginine
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: NWG, MG, PEG, PGE, EDO, NA |
 |
Crystal Structure Of Alpha Keto Acid C-Methyl-Transferases Mrsa Bound To Sam
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, SAM, NA, PEG, EDO |
 |
Crystal Structure Of Selenomethionine Derivatized Alpha Keto Acid C-Methyl-Transferases Mrsa
Organism: Pseudomonas syringae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: MG, NA |
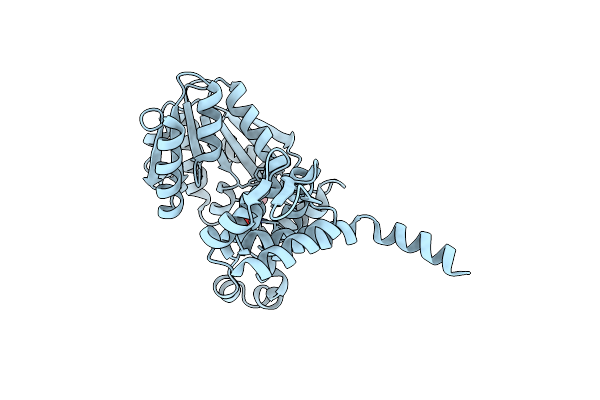 |
Crystal Structure Of Native Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL |
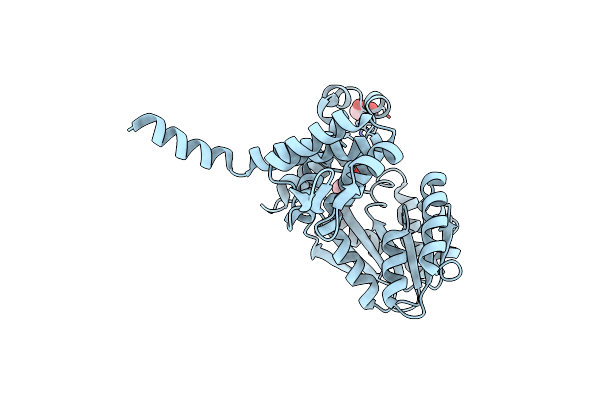 |
Crystal Structure Of S-Sad Phased Alpha-Keto C-Methyl Transferase Sgvm Bound To Ketoleucine
Organism: Streptomyces griseoviridis
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-07-03 Classification: TRANSFERASE Ligands: ZN, COI, CL, GOL |
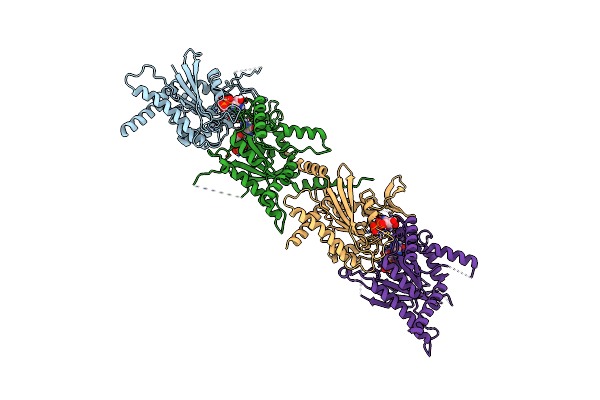 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2023-12-06 Classification: STRUCTURAL PROTEIN Ligands: GDP |

