Search Count: 46
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-07-22 Classification: ELECTRON TRANSPORT Ligands: ACE, HEC, LVQ |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: HEC, LVT, NA, SO4 |
 |
Organism: Salmonella enterica subsp. enterica serovar livingstone
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2016-06-29 Classification: UNKNOWN FUNCTION Ligands: HEM, GOL, NA, CL, MG |
 |
The Experimental X-Ray Structure Of The New Diheme Cytochrome Type C From Shewanella Baltica Os155 Sb-Dhc
Organism: Shewanella baltica
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-10-31 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Merohedral Twinning In Protein Crystals Revealed A New Synthetic Three Helix Bundle Motif
Organism: Synthetic
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-09-26 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Hiv-1 Protease - Epoxydic Inhibitor Complex (Ph 6 - Orthorombic Crystal Form P212121)
Organism: Human immunodeficiency virus type 1 (bru isolate)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2012-08-15 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 076, ACT, DMS |
 |
Hiv-1 Protease - Epoxydic Inhibitor Complex (Ph 9 - Monoclinic Crystal Form P21)
Organism: Human immunodeficiency virus type 1 (bru isolate)
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2012-08-15 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMS, 079 |
 |
Hiv-1 Protease - Epoxydic Inhibitor Complex (Ph 9 - Orthorombic Crystal Form P212121)
Organism: Human immunodeficiency virus type 1 (bru isolate)
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2012-08-15 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 079, DMS |
 |
Electron Transfer Complexes: Experimental Mapping Of The Redox-Dependent Cytochrome C Electrostatic Surface
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-01-25 Classification: ELECTRON TRANSPORT Ligands: HEC, NO3 |
 |
Electron Transfer Complexes:Experimental Mapping Of The Redox-Dependent Cytochrome C Electrostatic Surface
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-25 Classification: ELECTRON TRANSPORT Ligands: HEC, NO3 |
 |
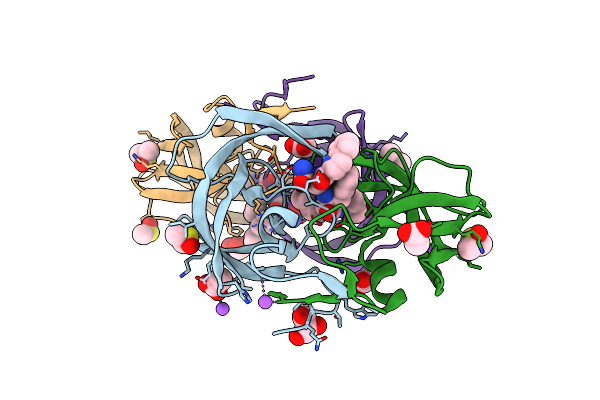
Organism: Human immunodeficiency virus type 1 bh10
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2011-07-20 Classification: hydrolase/hydrolase inhibitor Ligands: ROC, CL, DMS, NA |
 |
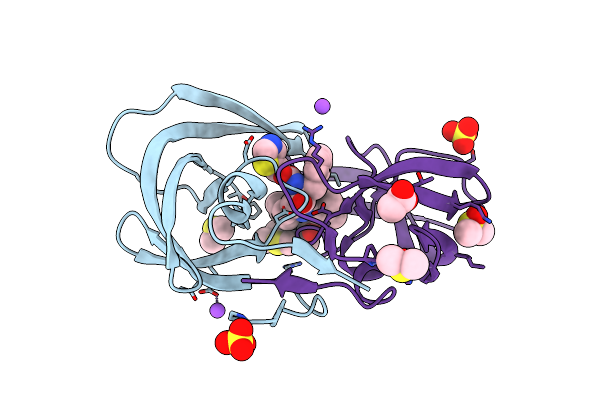
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2011-07-20 Classification: hydrolase/hydrolase inhibitor Ligands: ROC, DMS, ACT, CL, GOL, NA |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2011-07-20 Classification: hydrolase/hydrolase inhibitor Ligands: RIT, DMS, SO4, NA, ACT |
 |
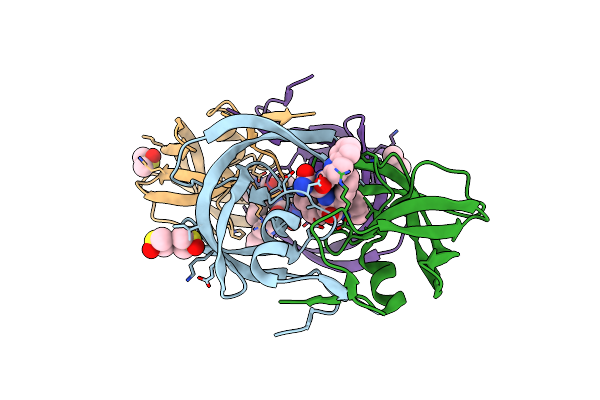
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2011-07-20 Classification: hydrolase/hydrolase inhibitor Ligands: RIT, DMS, GOL, CL, SO4 |
 |
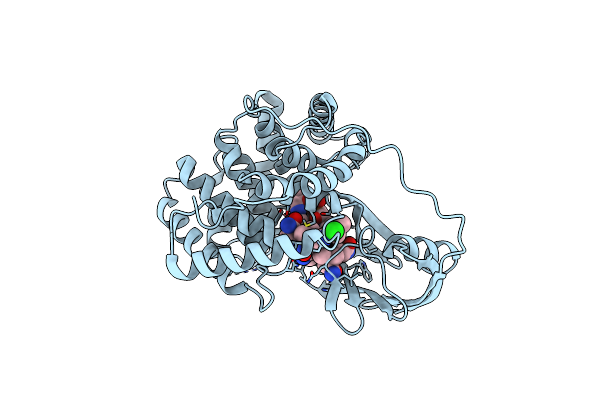
New Crystal Form Of Hiv-1 Protease/Saquinavir Structure Reveals Carbamylation Of N-Terminal Proline
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2010-06-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: DMS, ROC, CL |
 |
Crystallographic Analysis Of Upper Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CYN, CL |
 |
Crystallographic Analysis Of Beta-Axial Ligand Substitutions In Cobalamin Bound To Transcobalamin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-30 Classification: TRANSPORT PROTEIN Ligands: B12, CL, SO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: B12 |
 |
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Monoclinic Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |
 |
Structure Of Cobalamin-Complexed Bovine Transcobalamin In Trigonal Crystal Form
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-04-04 Classification: TRANSPORT PROTEIN Ligands: CL, B12 |

