Search Count: 53
 |
Organism: Francisella tularensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: APOPTOSIS Ligands: IOD |
 |
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: SIGNALING PROTEIN |
 |
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: SIGNALING PROTEIN |
 |
Insulin Receptor In Complex With Both Insulin And De Novo Designed Site-2 Binder "S2B".
Organism: Mus musculus, Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: SIGNALING PROTEIN |
 |
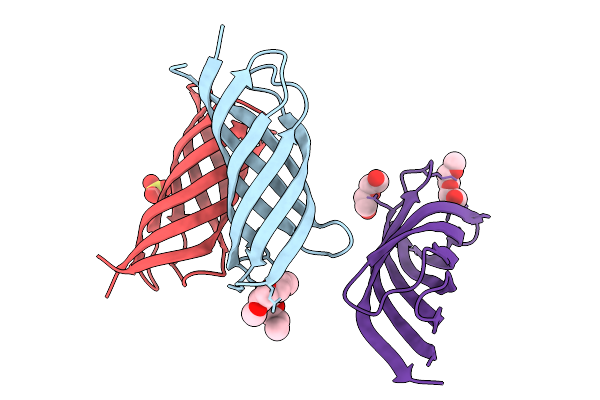
Structure Of A De Novo Designed Interleukin-21 Mimetic Complex With Il-21R And Il-2Rg
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: DE NOVO PROTEIN Ligands: PG6, SO4, PG4 |
 |
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: PROTEIN BINDING Ligands: CL, PGE, MES, TRS |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: APOPTOSIS |
 |
Organism: Acinetobacter terrae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: PG4 |
 |
Organism: Acinetobacter terrae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: MG, BTB |
 |
Designed Miniproteins Potently Inhibit And Protect Against Mers-Cov. Crystal Structure Of Mers-Cov S Rbd In Complex With Miniprotein Cb3
Organism: Middle east respiratory syndrome-related coronavirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: NAG |
 |
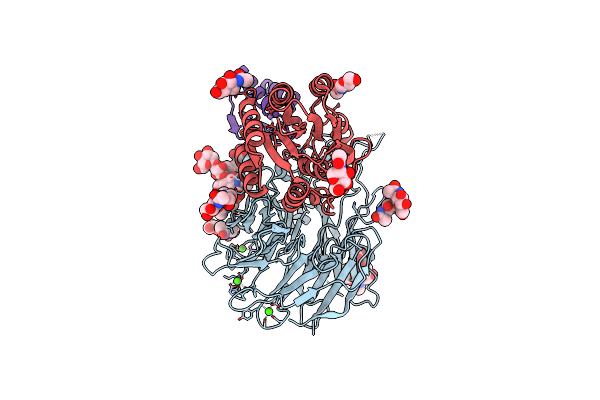
Cryo-Em Structure Of Alpha5Beta1 Integrin In Complex With Neonectin Candidate 2
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: CA, NAG, MN |
 |
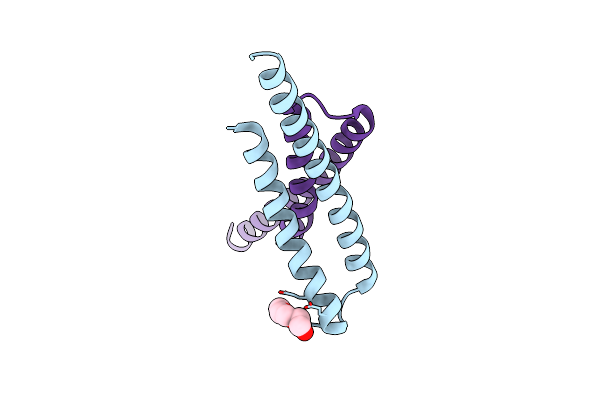
Cryo-Em Structure Of Alpha5Beta1 Integrin In Complex With Neonectin Candidate 2, Open Conformation
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: CA, NAG, MN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: DE NOVO PROTEIN Ligands: PG4 |
 |
De Novo Design Of High-Affinity Protein Binders To Bio Active Peptide Amylin
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: DE NOVO PROTEIN |
 |
De Novo Design Of High-Affinity Protein Binders To Rna Binding Domain Of G3Bp1
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: DE NOVO PROTEIN Ligands: SO4 |

