Search Count: 18
 |
Structure Of The Saccharomyces Cerevisiae Pcna Clamp Unloader Elg1-Rfc Complex
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Structure Of The Saccharomyces Cerevisiae Clamp Unloader Elg1-Rfc Bound To A Cracked Pcna
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Structure Of The Saccharomyces Cerevisiae Clamp Unloader Elg1-Rfc Bound To Pcna
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: REPLICATION Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 1 (Open 9-1-1 And Shoulder Bound Dna Only)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: MG, AGS, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 2 (Open 9-1-1 Ring And Flexibly Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 3 (Open 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 4 (Partially Closed 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 10-Nt Gapped Dna In Step 5 (Closed 9-1-1 And Stably Bound Chamber Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: MG, AGS, ADP |
 |
Structure Of S. Cerevisiae Rad24-Rfc Loading The 9-1-1 Clamp Onto A 5-Nt Gapped Dna (9-1-1 Encircling Fully Bound Dna)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: CELL CYCLE/DNA Ligands: AGS, MG, ADP |
 |
Structure Of The Yeast Rad24-Rfc Loader Bound To Dna And The Closed 9-1-1 Clamp
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: DNA BINDING PROTEIN/DNA Ligands: AGS, MG, ADP, PO4 |
 |
Structure Of The Yeast Rad24-Rfc Loader Bound To Dna And The Open 9-1-1 Clamp
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: DNA BINDING PROTEIN/DNA Ligands: AGS, MG, ADP |
 |
Structure Of The Saccharomyces Cerevisiae Replicative Polymerase Delta In Complex With A Primer/Template And The Pcna Clamp
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-12-02 Classification: REPLICATION Ligands: MG, SF4, D3T, ZN |
 |
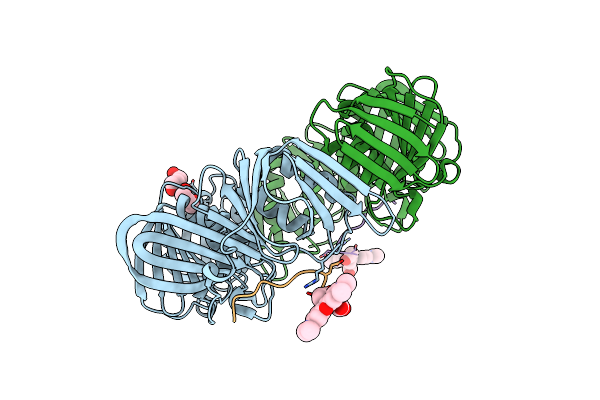
Crystal Structure Of E. Coli Sliding Clamp (Beta) Bound To A Polymerase Ii Peptide
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-07-29 Classification: TRANSFERASE, TRANSCRIPTION |
 |
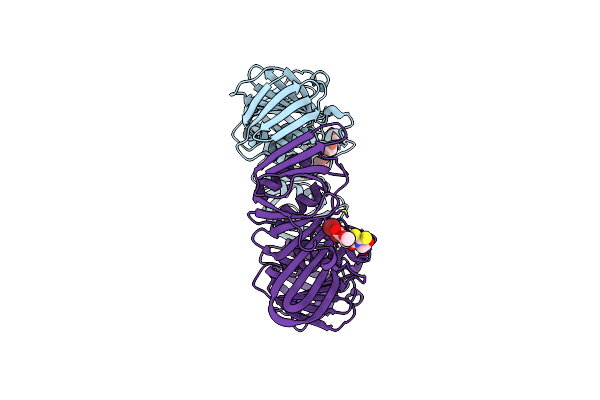
Crystal Structure Of E. Coli Sliding Clamp (Beta) Bound To A Polymerase Iii Peptide
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-07-29 Classification: TRANSFERASE, TRANSCRIPTION Ligands: PEG, 323 |
 |
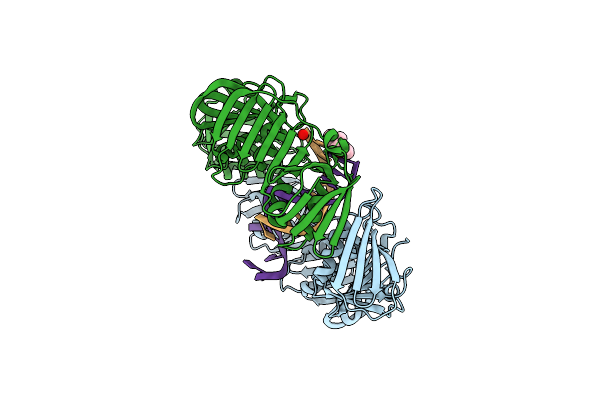
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2008-07-29 Classification: TRANSFERASE, TRANSCRIPTION Ligands: 322 |
 |
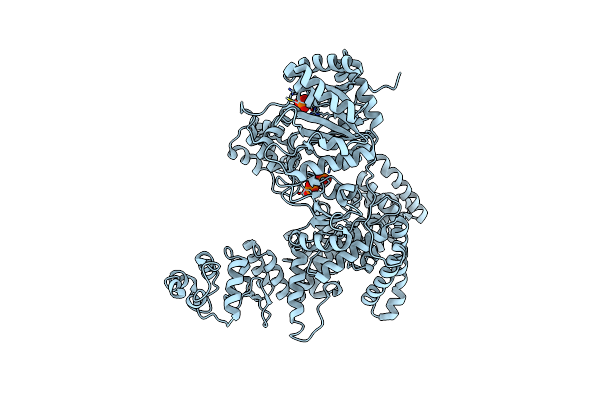
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2008-01-29 Classification: TRANSFERASE, TRANSCRIPTION/DNA Ligands: 5CY |
 |
Crystal Structure Of The Catalytic Alpha Subunit Of E. Coli Replicative Dna Polymerase Iii
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-09-19 Classification: TRANSFERASE Ligands: PO4 |
 |
Crystal Structure Of The Catalytic Alpha Subunit Of E. Coli Replicative Dna Polymerase Iii
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-09-19 Classification: TRANSFERASE Ligands: PO4 |

