Search Count: 18
 |
Organism: Photinus pyralis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSLATION Ligands: MG, ZN |
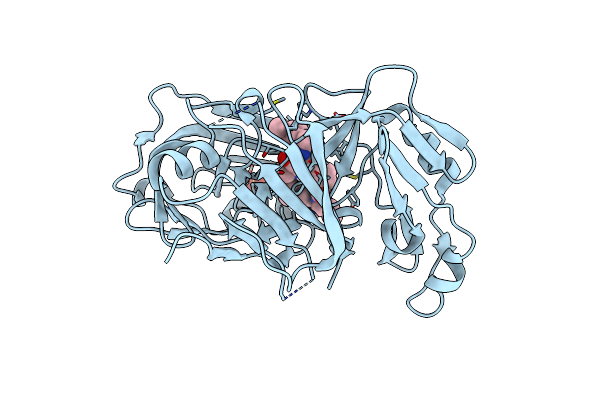 |
Crystal Structure Of Plasmodium Falciparum Plasmepsin X In Complex With The Hydroxyethylamine Drug 7K.
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: A1ITU, SO4 |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: REPLICATION Ligands: ZN |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: REPLICATION Ligands: ATP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2021-06-09 Classification: TRANSCRIPTION Ligands: SO4, GOL, TAR |
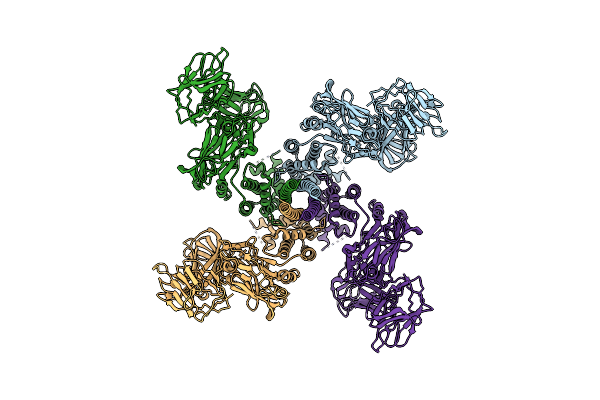 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2021-04-14 Classification: TOXIN |
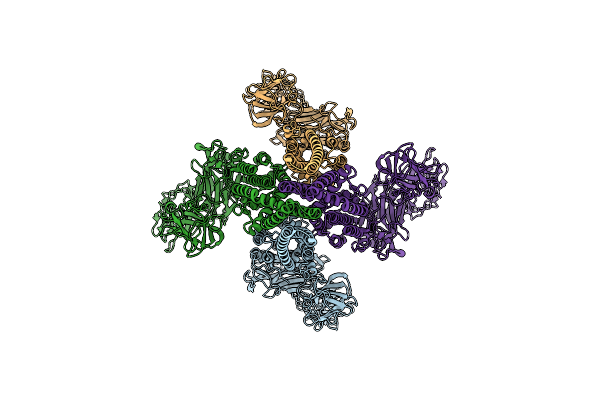 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: TOXIN |
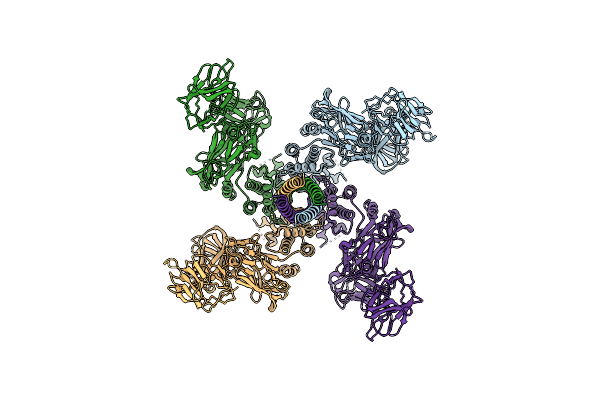 |
Organism: Bacillus thuringiensis
Method: ELECTRON MICROSCOPY Resolution:4.80 Å Release Date: 2021-03-17 Classification: TOXIN |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-08-07 Classification: CELL CYCLE |
 |
Organism: Schizosaccharomyces pombe, Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-08-07 Classification: CELL CYCLE Ligands: BR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-06-05 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Ret Recognition Of Gdnf-Gfralpha1 Ligand By A Composite Binding Site Promotes Membrane-Proximal Self-Association
Organism: Homo sapiens, Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:24.00 Å Release Date: 2014-10-01 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-05-29 Classification: SIGNALING PROTEIN Ligands: PE4, FAR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-12-08 Classification: IMMUNE SYSTEM Ligands: NAG, GOL |
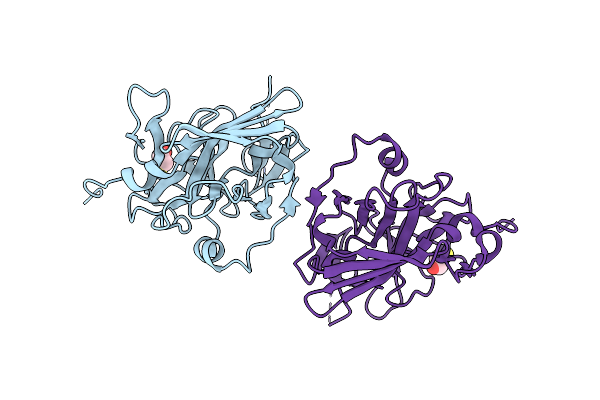 |
Organism: Tobacco etch virus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-11-02 Classification: Viral protein, hydrolase Ligands: BME |
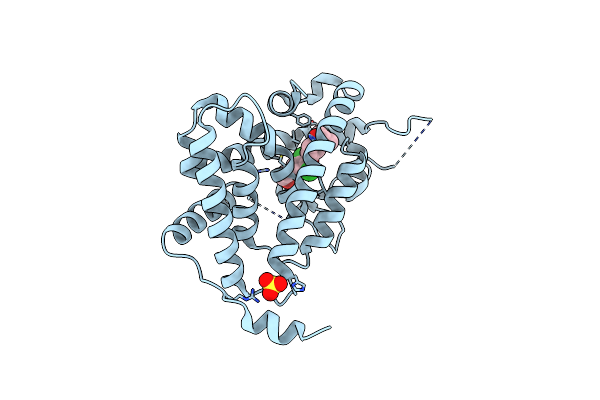 |
Thyroid Receptor Alpha In Complex With An Agonist Selective For Thyroid Receptor Beta1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-06-17 Classification: MEMBRANE PROTEIN Ligands: SO4, IH5 |
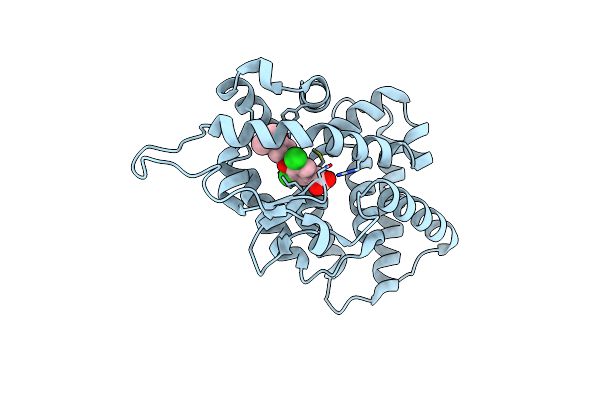 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2003-06-17 Classification: MEMBRANE PROTEIN Ligands: IH5 |

