Search Count: 11
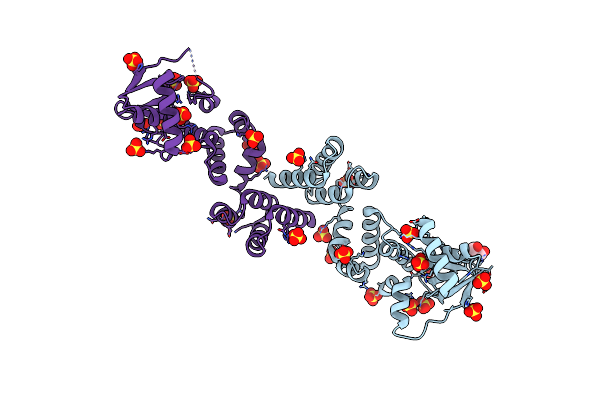 |
Crystal Structure Of C-Terminally Truncated Geobacillus Thermoleovorans Nucleoid Occlusion Protein Noc
Organism: Geobacillus thermoleovorans ccb_us3_uf5
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-02-17 Classification: DNA BINDING PROTEIN Ligands: SO4, GOL |
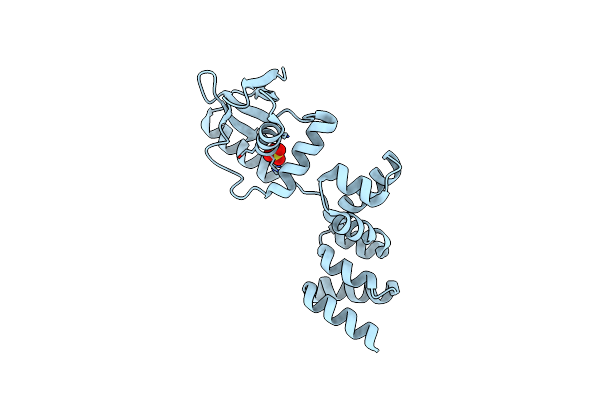 |
Crystal Structure Of N- And C-Terminally Truncated Geobacillus Thermoleovorans Nucleoid Occlusion Protein Noc
Organism: Geobacillus thermoleovorans ccb_us3_uf5
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-02-17 Classification: DNA BINDING PROTEIN Ligands: SO4 |
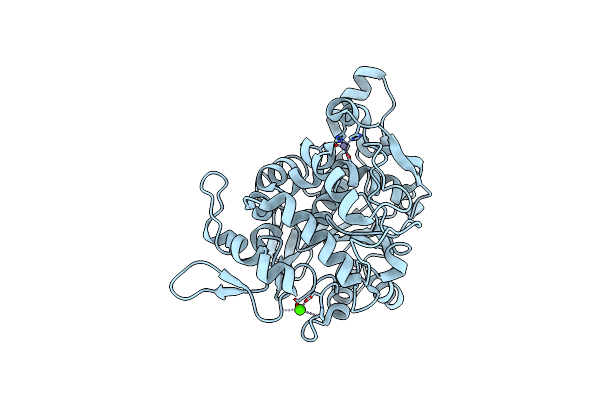 |
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: CA, ZN |
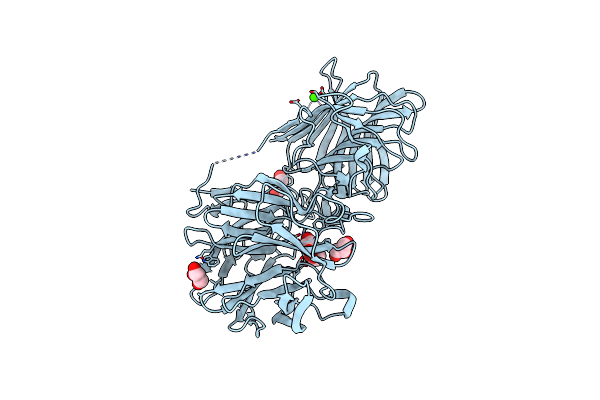 |
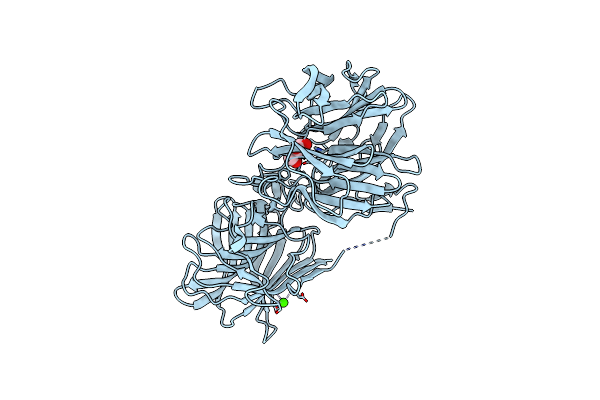
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, GOL |
 |
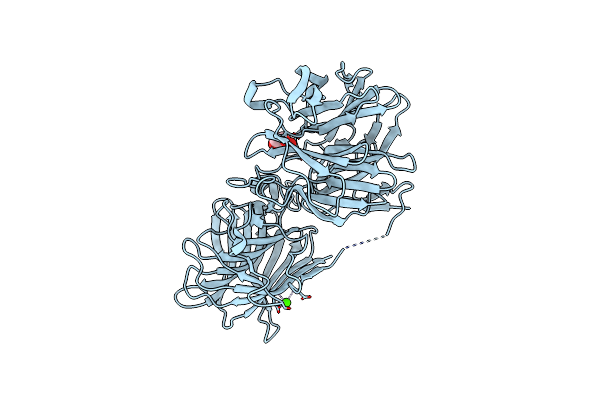
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With L-Arabinose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, FUB |
 |
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With D-Xylose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, XYS |
 |
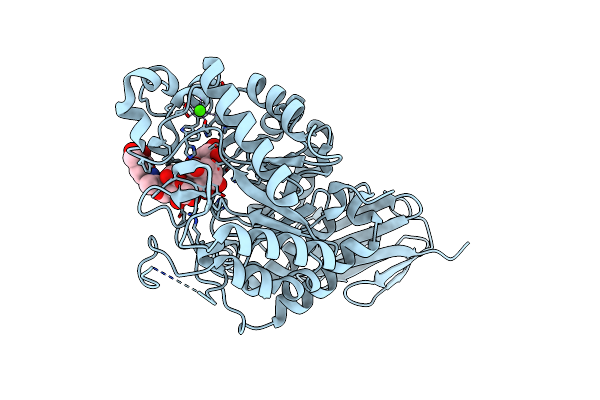
Crystal Structure Of A Thermostable Glycoside Hydrolase Family 43 {Beta}-1,4-Xylosidase From Geobacillus Thermoleovorans It-08 In Complex With L-Arabinose And D-Xylose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-04-25 Classification: HYDROLASE Ligands: CA, FUB, XYS, XYP |
 |
Crystal Structure Of Alpha-Amylase From Geobacillus Thermoleovorans, Gta, Complexed With Acarbose
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-03-13 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, ACI |
 |
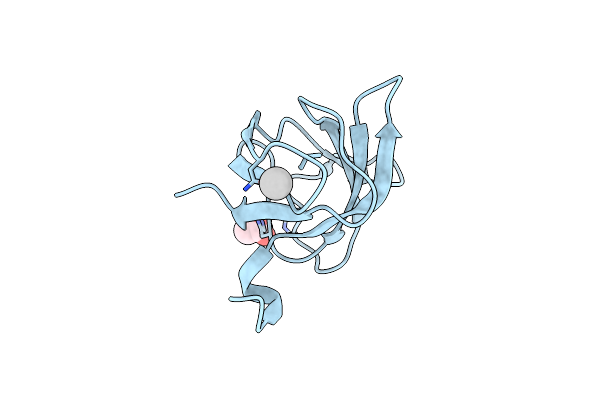
Crystal Structure Of The Selenomethionine Variant Of The C-Terminal Domain Of Geobacillus Thermoleovorans Putative U32 Peptidase
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-11-14 Classification: UNKNOWN FUNCTION Ligands: SO4 |
 |
Crystal Structure Of The C-Terminal Domain Of Geobacillus Thermoleovorans Putative U32 Peptidase
Organism: Geobacillus thermoleovorans
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-14 Classification: UNKNOWN FUNCTION Ligands: UNX, ACT |
 |
NA
Organism: Geobacillus thermoleovorans
Method: Alphafold Release Date: Classification: NA Ligands: NA |

