Search Count: 25
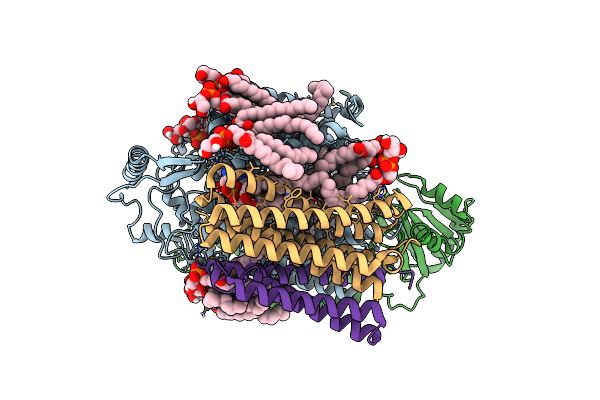 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.19 Å Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN Ligands: UQ8, LHG, 3PE, HEO, HEM, CDL, CU, ZN |
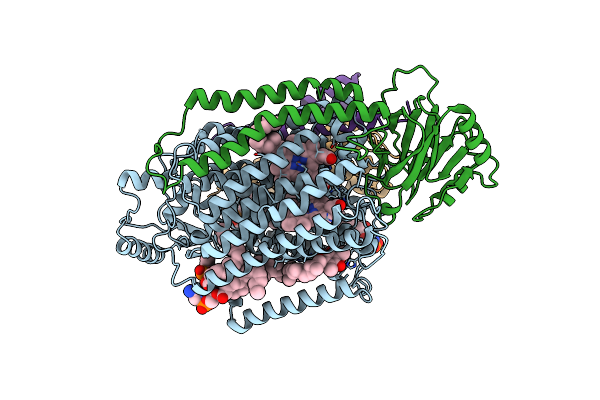 |
2.55-Angstrom Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli In Native Membrane
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: OXIDOREDUCTASE Ligands: HEM, HEO, CU, 3PE, UQ8 |
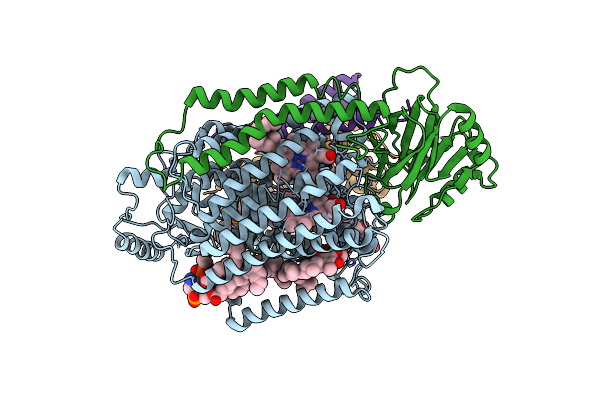 |
2.55-Angstrom Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli In Native Membrane
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: OXIDOREDUCTASE Ligands: HEM, HEO, CU, 3PE, UQ8 |
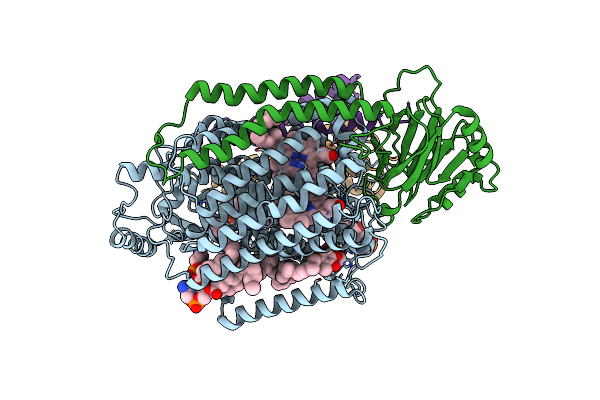 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2021-08-25 Classification: OXIDOREDUCTASE Ligands: HEM, HEO, CU, 3PE, UQ8 |
 |
X-Ray Structure Of The Proton-Pumping Cytochrome Aa3-600 Menaquinol Oxidase From Bacillus Subtilis
Organism: Bacillus subtilis, Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MQ7 |
 |
X-Ray Structure Of The Proton-Pumping Cytochrome Aa3-600 Menaquinol Oxidase From Bacillus Subtilis Complexed With 3-Iodo-N-Oxo-2-Heptyl-4-Hydroxyquinoline
Organism: Bacillus subtilis, Bacillus subtilis subsp. subtilis str. 168, Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: HEA, CU, IHQ |
 |
X-Ray Structure Of The Proton-Pumping Cytochrome Aa3-600 Menaquinol Oxidase From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2020-01-15 Classification: OXIDOREDUCTASE Ligands: HEA, CU, HQO |
 |
Cryo-Em Structure Of The E. Coli Cytochrome Bd-I Oxidase At 2.68 A Resolution
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2019-10-16 Classification: MEMBRANE PROTEIN Ligands: UQ8, POV, HDD, HEB, OXY |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, COA |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, COA |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-09-25 Classification: OXIDOREDUCTASE Ligands: FAD, COA, VK3 |
 |
Structure Of Alternative Complex Iii From Flavobacterium Johnsoniae (Wild Type)
Organism: Flavobacterium johnsoniae uw101
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2018-05-09 Classification: MEMBRANE PROTEIN Ligands: HEC, F3S, SF4, DKA, FAW |
 |
Critical Role Of Water Molecules For Proton Translocation Of The Membrane-Bound Transhydrogenase
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-10 Classification: OXIDOREDUCTASE Ligands: 1PE, PEG, OLC, BEN |
 |
R. Sphaeroides Photosythetic Reaction Center Mutant - Residue L223, Ser To Trp - Room Temperature Structure Solved On X-Ray Transparent Microfluidic Chip
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2017-04-12 Classification: PHOTOSYNTHESIS Ligands: BPH, BCL, FE, U10 |
 |
Organism: Nitrosopumilus maritimus (strain scm1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-05-11 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Mechanism Of Transhydrogenase Coupling Proton Translocation And Hydride Transfer
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:6.93 Å Release Date: 2015-01-28 Classification: MEMBRANE PROTEIN Ligands: NAP, NAD |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-12-31 Classification: MEMBRANE PROTEIN Ligands: HG |
 |
Crystal Structure Of Thermus Thermophilis Transhydrogeanse Domain Ii Dimer Semet Derivative
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2014-06-11 Classification: MEMBRANE PROTEIN |
 |
Mechanism Of Transhydrogenase Coupling Proton Translocation And Hydride Transfer
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2014-06-11 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Alpha1 Dimer Of Thermus Thermophilus Transhydrogenase In P4(3)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-02-05 Classification: OXIDOREDUCTASE Ligands: GOL |

