Search Count: 34
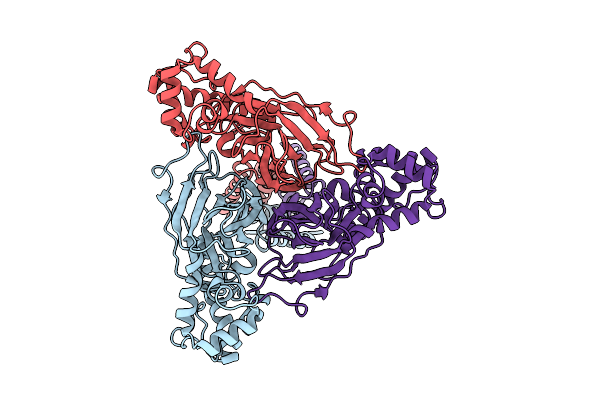 |
Organism: Scolopendra mutilans
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: SIGNALING PROTEIN |
 |
Structure Of Xenopus Tropicalis Acid-Sensitive Outwardly Rectifying Channel Asor Trimer Bound With Trna (Intermediate State)
Organism: Xenopus tropicalis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Dimeric Xenopus Tropical Acid-Sensitive Outwardly Rectifying Channel Asor Trimer Bound With Trna (Closed State)
Organism: Xenopus tropicalis, Spodoptera frugiperda
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN |
 |
Structure Of Xenopus Tropicalis Acid-Sensitive Outwardly Rectifying Channel Asor (Resting State)
Organism: Xenopus tropicalis
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN |
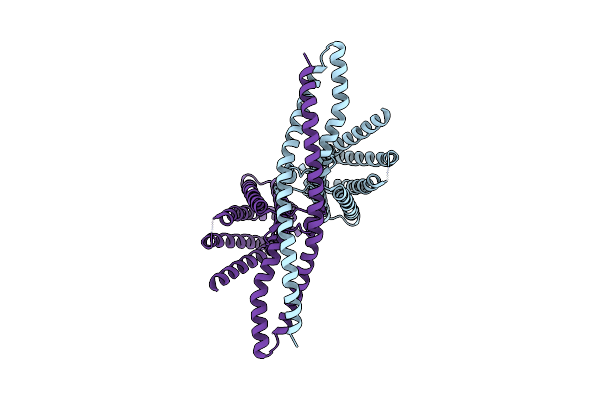 |
Organism: Anaplasma marginale, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN |
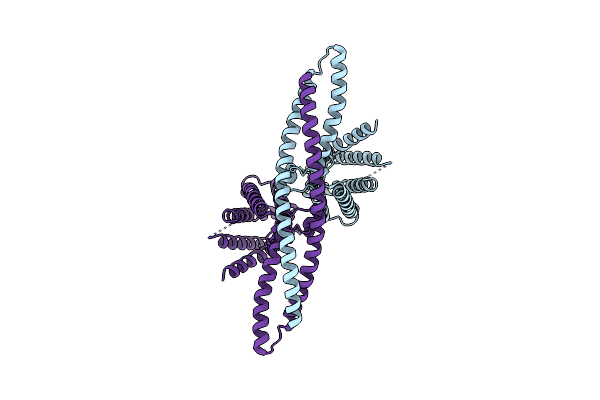 |
Organism: Anaplasma marginale, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-09-01 Classification: MEMBRANE PROTEIN |
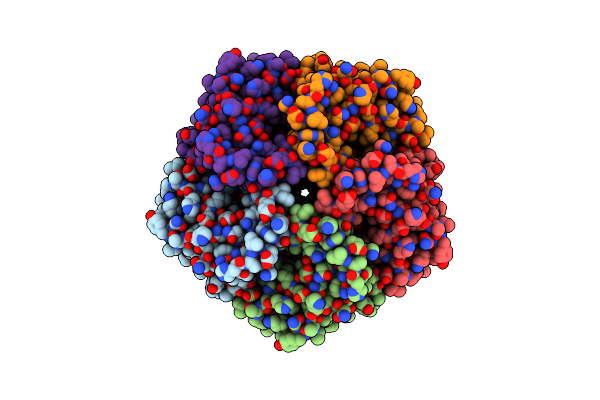 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSPORT PROTEIN Ligands: HV6 |
 |
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE |
 |
1.98 A Resolution Crystal Structure Of Group A Streptococcus H111A Hupz-V5-His6
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
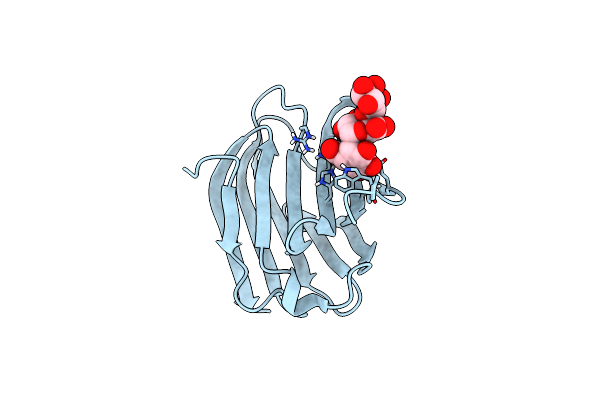
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2020-08-26 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-08-26 Classification: SUGAR BINDING PROTEIN |
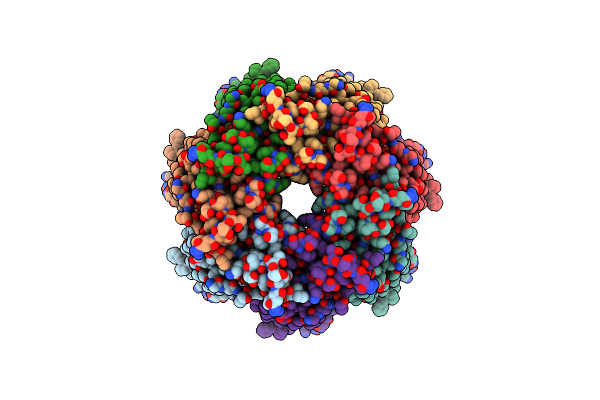 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: TRANSPORT PROTEIN |
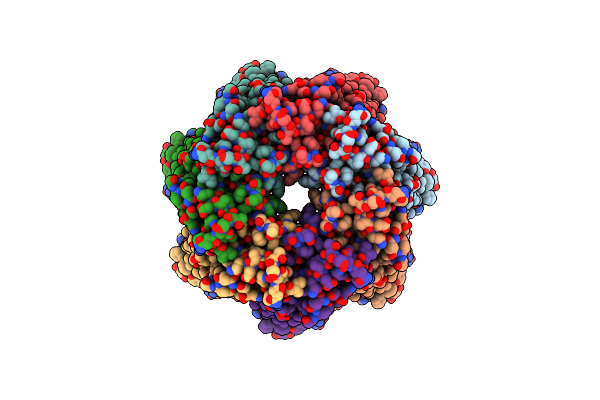 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: TRANSPORT PROTEIN |
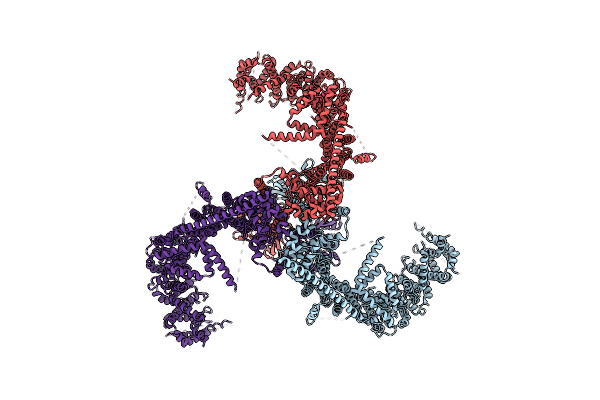 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2020-03-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-11-21 Classification: TRANSPORT PROTEIN Ligands: BNG |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2018-01-31 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2016-08-31 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5BS, CL, SO4 |
 |
1.5 Angstrom Crystal Structure Of 3-Hydroxyanthranilate-3,4-Dioxygenase From Cupriavidus Metallidurans
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2016-03-02 Classification: OXIDOREDUCTASE Ligands: FE2, TRS |

