Search Count: 26
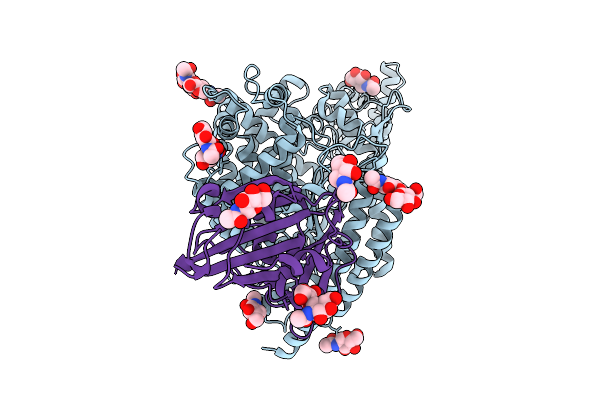 |
Structure Of The Rattus Norvegicus Ace2 Receptor Bound Hsitaly2011 Rbd Complex
Organism: Rattus norvegicus, Merbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG, ZN |
 |
Organism: Eptesicus fuscus, Merbecovirus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE/VIRAL PROTEIN Ligands: NAG, ZN |
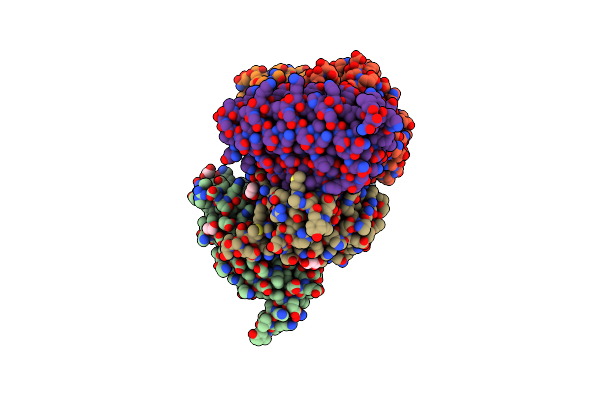 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SIGNALING PROTEIN Ligands: EDO, MG, GNP, 1PE, PGE, SRT |
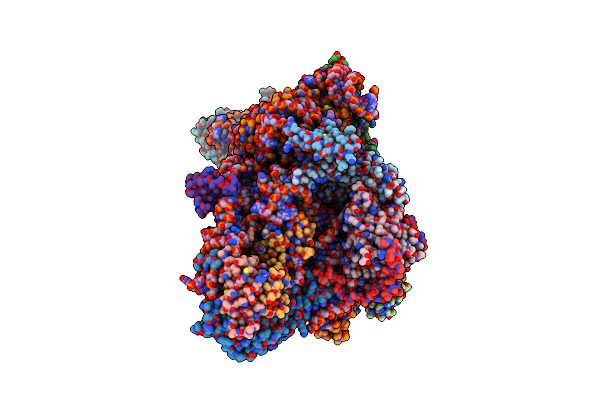 |
Sars-Cov-2 Nsp1 Bound To The Rhinolophus Lepidus 40S Ribosomal Subunit (Local Refinement Of The 40S Body)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens, Rhinolophus lepidus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: RIBOSOME Ligands: MG, K, ZN |
 |
Sars-Cov-2 Nsp1 Bound To The Rhinolophus Lepidus 40S Ribosome (Local Refinement Of The 40S Head)
Organism: Rhinolophus lepidus
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: RIBOSOME Ligands: MG, ZN, K |
 |
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2025-02-26 Classification: VIRAL PROTEIN Ligands: NAG, ZN, FOL, EIC |
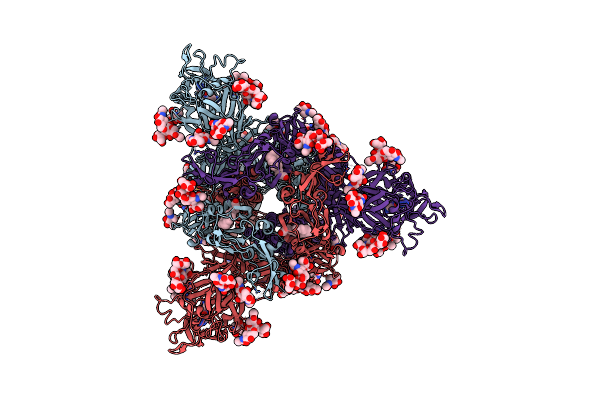 |
Organism: Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2025-02-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, FOL, EIC |
 |
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
Organism: Bos taurus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: SIGNALING PROTEIN |
 |
 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Rat Stat2 Ccd
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Full-Length Rat Stat2
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
 |
Organism: Arabidopsis thaliana
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-09 Classification: FLAVOPROTEIN, OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2019-01-09 Classification: FLAVOPROTEIN, OXIDOREDUCTASE Ligands: FMN, GOL |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-01-09 Classification: FLAVOPROTEIN, Oxidoreductase Ligands: GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2017-03-08 Classification: ENDOCYTOSIS Ligands: AGS, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.27 Å Release Date: 2017-03-08 Classification: ENDOCYTOSIS Ligands: ADP, MG |
 |
Crystal Structure Of A Trimeric Influenza Hemagglutinin Stem In Complex With An Broadly Neutralizing Antibody Cr9114
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-09-09 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

