Search Count: 8
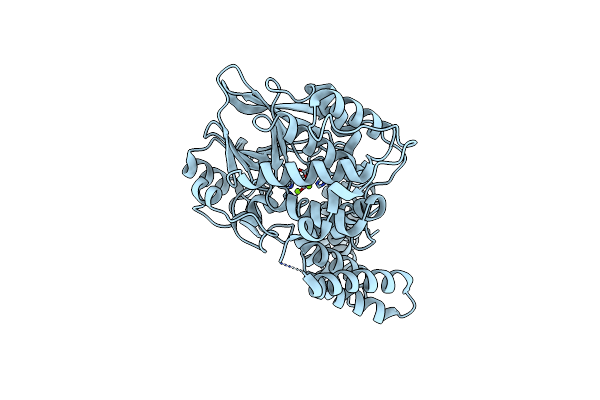 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: MG |
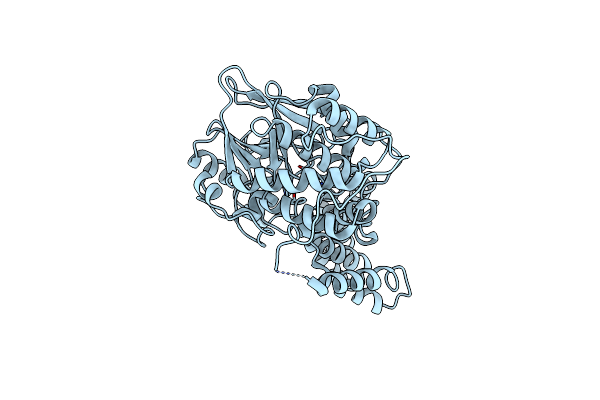 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-02-19 Classification: HYDROLASE Ligands: MG |
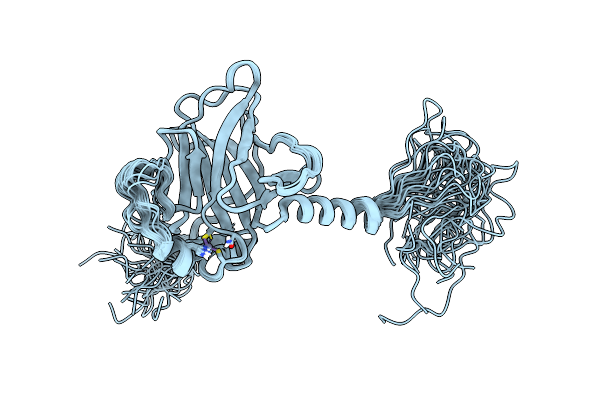 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2008-11-11 Classification: CELL CYCLE, ANTITUMOR PROTEIN Ligands: ZN |
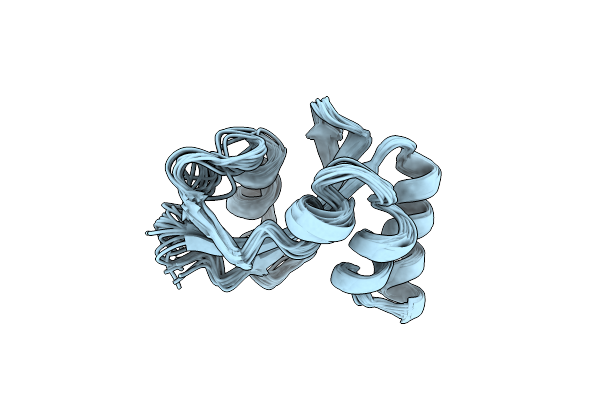 |
Nmr Structure Of The E. Coli Peptidyl-Prolyl Cis/Trans-Isomerase Parvulin 10
|
 |
Nmr Structure Of The E. Coli Peptidyl-Prolyl Cis/Trans-Isomerase Parvulin 10
|
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1996-10-14 Classification: PHOSPHOTRANSFERASE |
 |
Nmr Studies Of (U-13C)Cyclosporin A Bound To Cyclophilin: Bound Conformation And Portions Of Cyclosporin Involved In Binding
Organism: Tolypocladium inflatum
Method: SOLUTION NMR Release Date: 1994-01-31 Classification: IMMUNOSUPPRESSANT |
 |
Nmr Studies Of (U-13C)Cyclosporin A Bound To Cyclophilin: Bound Conformation And Portions Of Cyclosporin Involved In Binding
Organism: Tolypocladium inflatum
Method: SOLUTION NMR Release Date: 1994-01-31 Classification: IMMUNOSUPPRESSANT |

