Search Count: 13
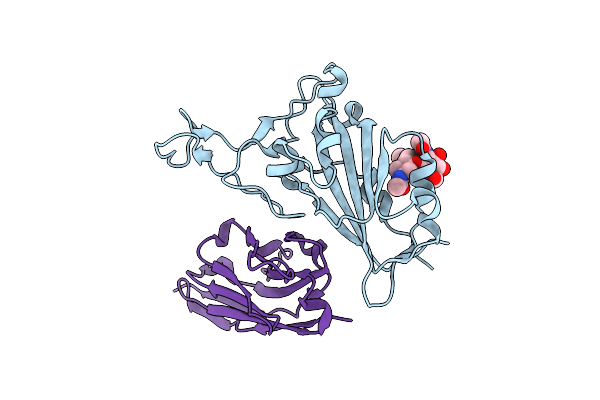 |
Crystal Structure Of The Sars-Cov-2 Neutralizing Vhh 7A9 Bound To The Spike Receptor Binding Domain
Organism: Severe acute respiratory syndrome coronavirus 2, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-08-16 Classification: ANTIVIRAL PROTEIN |
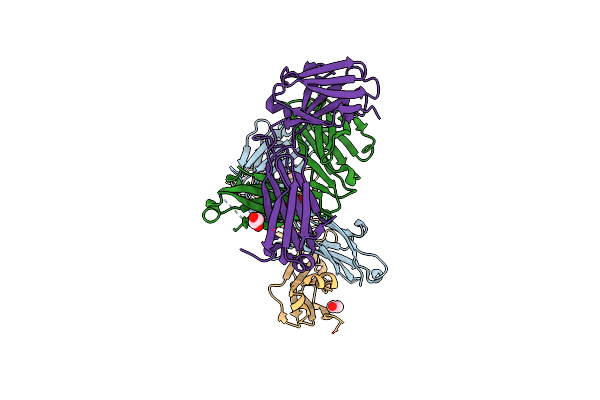 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-02-14 Classification: IMMUNE SYSTEM Ligands: EDO |
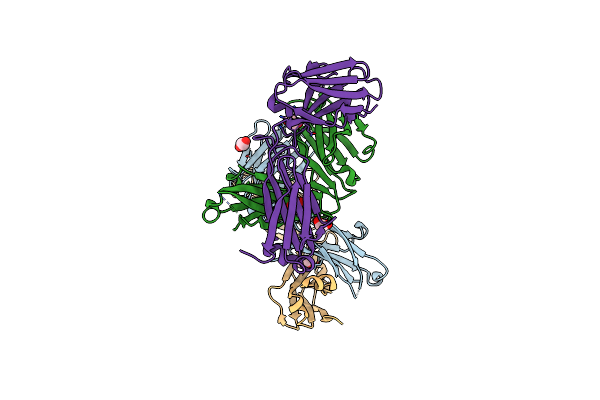 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2018-02-14 Classification: IMMUNE SYSTEM Ligands: EDO |
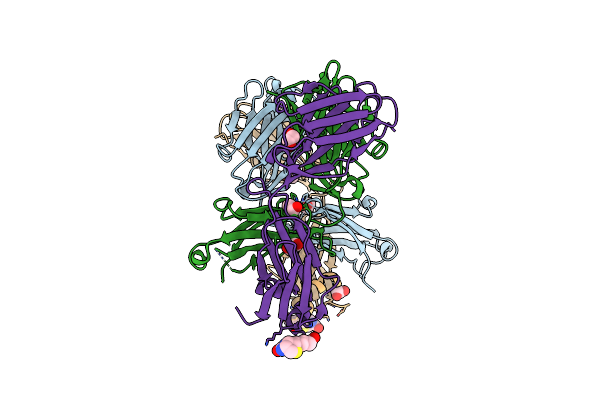 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-14 Classification: IMMUNE SYSTEM Ligands: EDO, BTN |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: MES |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of Ddn, The Deazaflavin-Dependent Nitroreductase From Mycobacterium Tuberculosis Involved In Bioreductive Activation Of Pa-824, With Co-Factor F420
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-01-18 Classification: OXIDOREDUCTASE Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Nitroreductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42 |
 |
Structure Of A Deazaflavin-Dependent Reductase From Nocardia Farcinica, With Co-Factor F420
Organism: Nocardia farcinica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-01-18 Classification: UNKNOWN FUNCTION Ligands: F42, SO4 |
 |
Efforts Toward Expansion Of The Genetic Alphabet: Structure And Replication Of Unnatural Base Pairs
Method: X-RAY DIFFRACTION
Resolution:2.80 Å Release Date: 2007-10-30 Classification: DNA Ligands: MG |
 |
Covalent Modification Of Guanine Bases In Double Stranded Dna: The 1.2 Angstroms Z-Dna Structure Of D(Cgcgcg) In The Presence Of Cucl2
Method: X-RAY DIFFRACTION
Resolution:1.20 Å Release Date: 1992-04-15 Classification: DNA Ligands: CU, NA |
 |
Base Specific Binding Of Copper(Ii) To Z-Dna: The 1.3-Angstroms Single Crystal Structure Of D(M5Cguam5Cg) In The Presence Of Cucl2
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 1992-04-15 Classification: DNA Ligands: CU, CUL |

