Search Count: 910
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: ZN, A1C09 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1C1A, CL, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: A1IR8 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: A1IR9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: A1JLU |
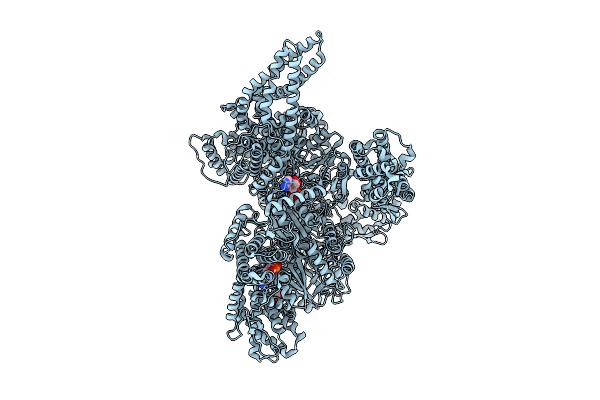 |
Motor Domain With Apo Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ATP, ADP, MG |
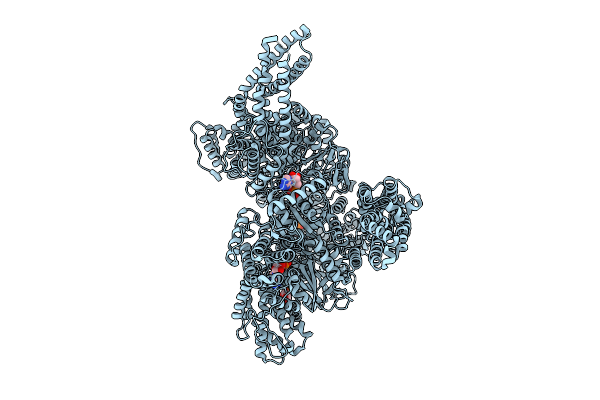 |
Motor Domain With Adp Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ADP, ATP |
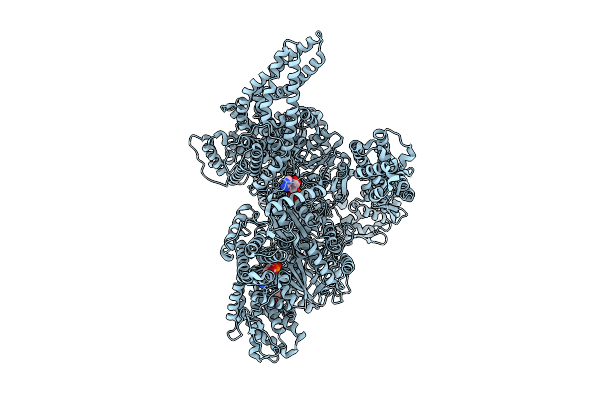 |
Motor Domain Alone With Apo Aaa1 And Adp Aaa3 From Yeast Full-Length Dynein-1 And Pac1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ATP, ADP, MG |
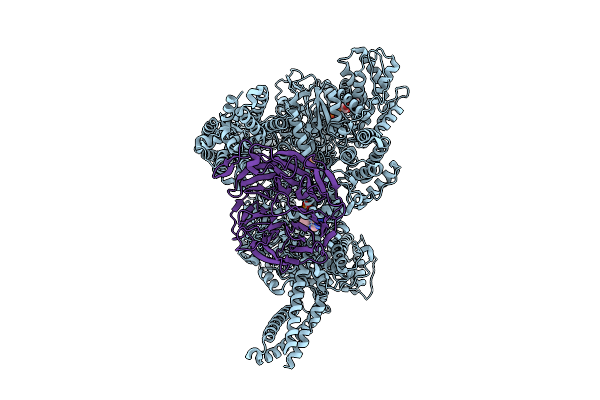 |
Motor Domain-Pac1 Complex With Adp Aaa1 And Apo Aaa3 From Yeast Full-Length Dynein-1 And Pac1 In 0.1 Mm Atp Condition
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-12-17 Classification: MOTOR PROTEIN Ligands: ADP, ATP, MG |
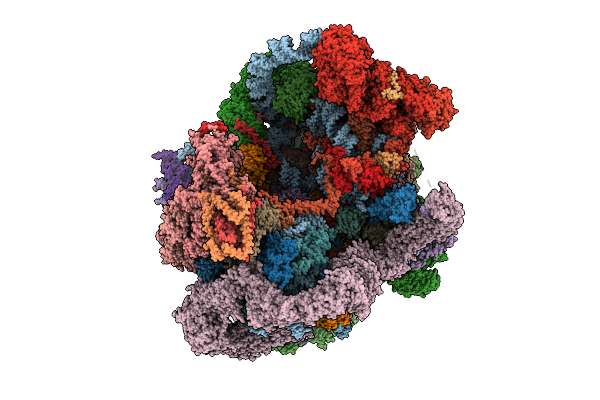 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
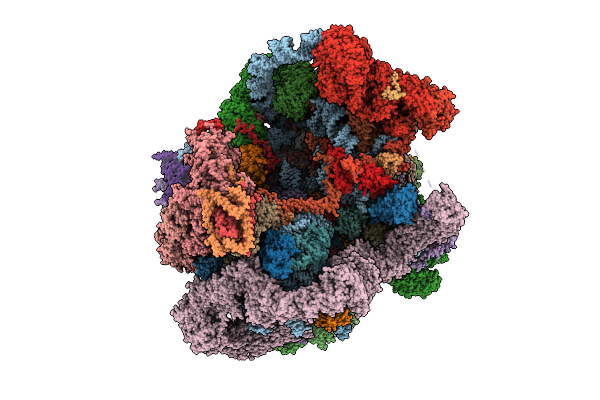 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
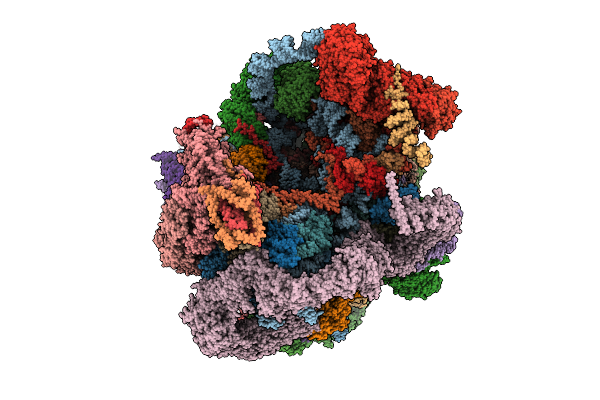 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
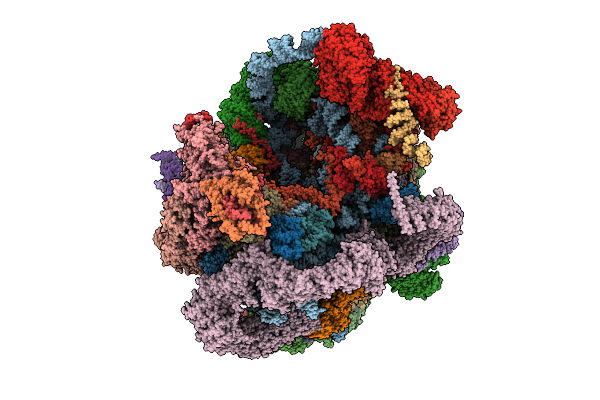 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, GTP |
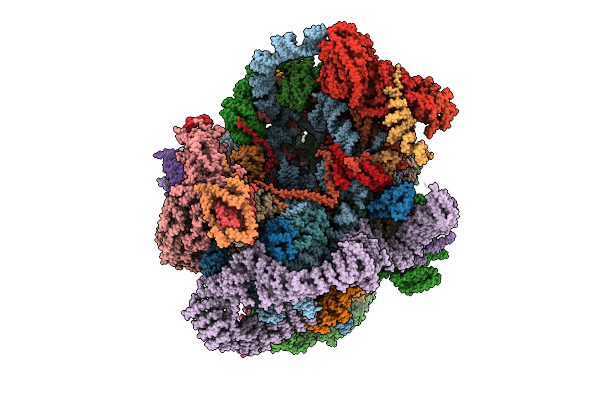 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
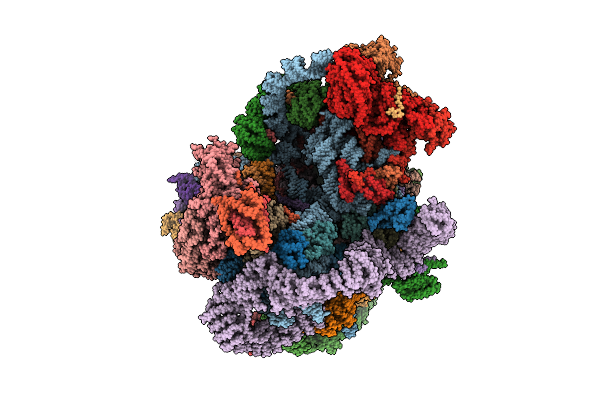 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
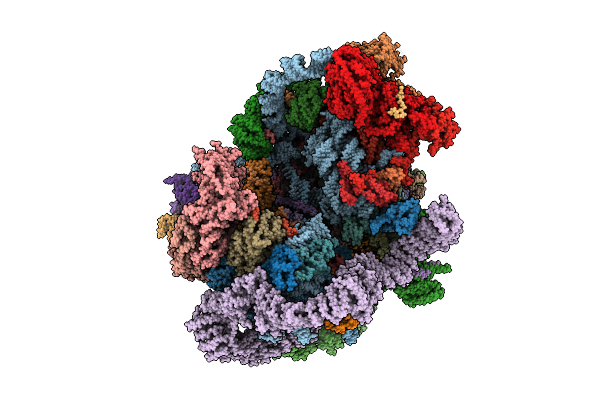 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME |
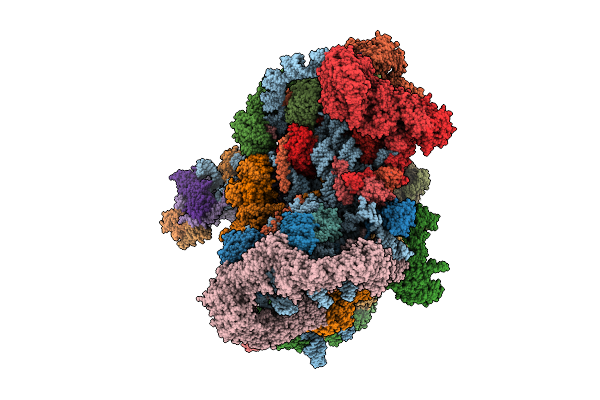 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, ADP, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ATP, ZN, ADP, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ZN, ADP, GTP |
 |
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RIBOSOME Ligands: MG, ZN, GTP |

