Search Count: 18
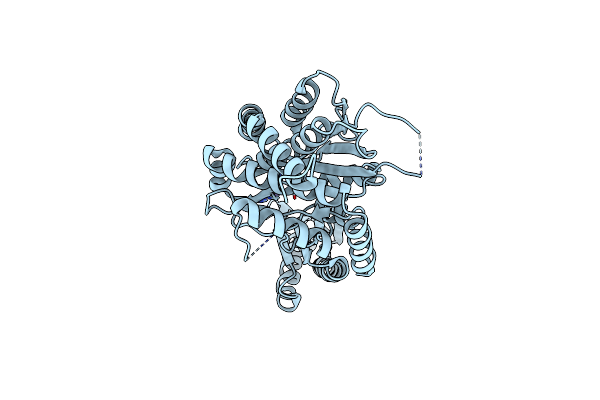 |
Crystal Structure Of Human Cytosolic Beta-Alanyl Lysine Dipeptidase (Pm20D2)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-08-02 Classification: HYDROLASE Ligands: ZN |
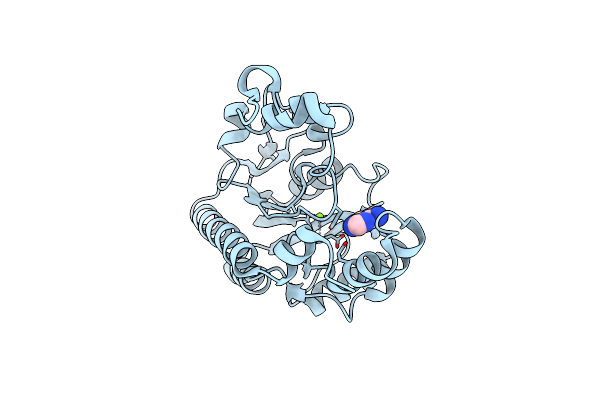 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenine At Ph 9
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADE, MG |
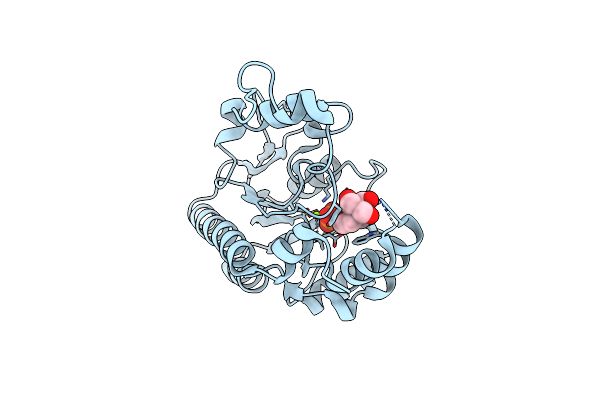 |
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Bis-Tris At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: BTB, MG, PO4 |
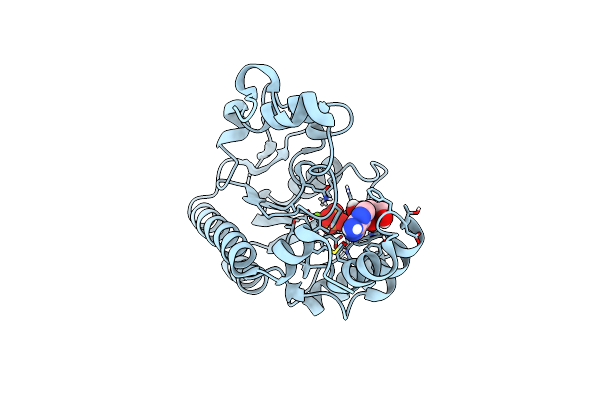 |
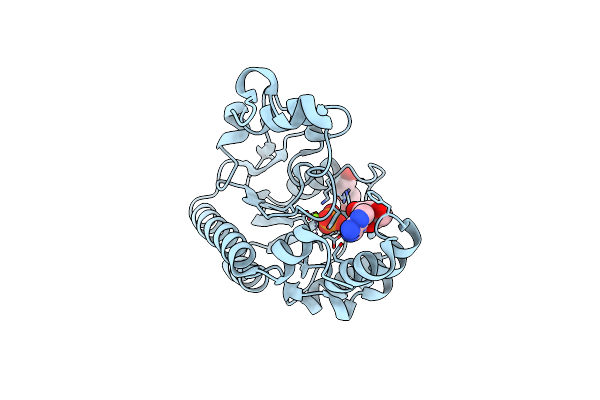
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenosine At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADN, PO4, MG |
 |
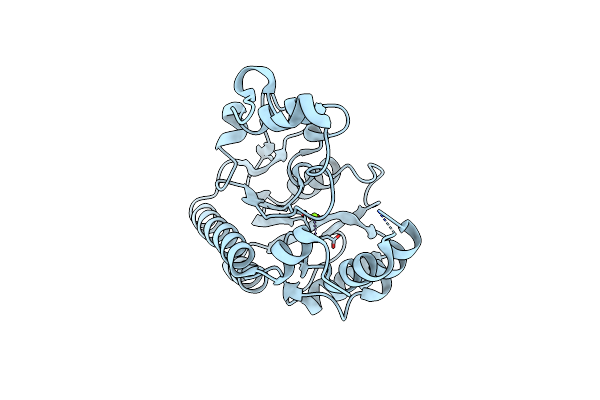
Crystal Structure Of Non-Specific Class-C Acid Phosphatase From Sphingobium Sp. Rsms Bound To Adenosine At Ph 5.5
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: ADN, PEG, MG, PO4 |
 |
Organism: Sphingobium sp. 20006fa
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2021-11-10 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of M1 Zinc Metallopeptidase E323A Mutant Bound To Tyr-Ser-Ala Substrate From Deinococcus Radiodurans
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422), Deinococcus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-25 Classification: HYDROLASE Ligands: ZN, FMT |
 |
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-07-17 Classification: HYDROLASE Ligands: ZN, TYR, NA |
 |
Crystal Structure Of S9 Peptidase (Inactive Form) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: ACT |
 |
Crystal Structure Of S9 Peptidase (Active Form) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE |
 |
Crystal Structure Of S9 Peptidase Mutant (S514A) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans (strain atcc 13939 / dsm 20539 / jcm 16871 / lmg 4051 / nbrc 15346 / ncimb 9279 / r1 / vkm b-1422)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: DMS, GOL |
 |
Crystal Structure Of S9 Peptidase (Inactive State)From Deinococcus Radiodurans R1 In P212121
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: GOL |
 |
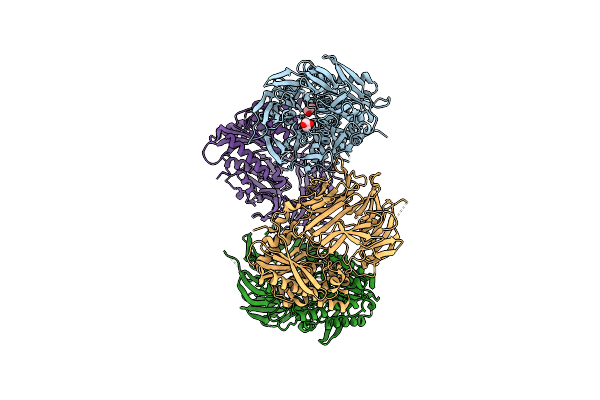
Crystal Structure Of Inactive State Of S9 Peptidase From Deinococcus Radiodurans R1 (Pmsf Treated)
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Crystal Structure Of S9 Peptidase (S514A Mutant In Inactive State) From Deinococcus Radiodurans R1
Organism: Deinococcus radiodurans str. r1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Substrate-Bound S9 Peptidase (S514A Mutant) From Deinococcus Radiodurans
Organism: Deinococcus radiodurans r1, Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: HYDROLASE Ligands: GOL |
 |
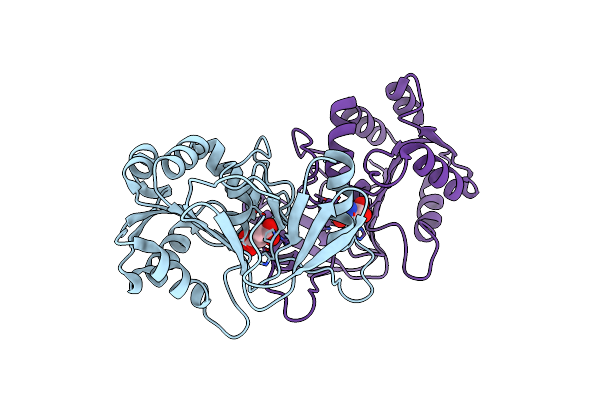
Crystal Structure Of Peptidase E With Ordered Active Site Loop From Salmonella Enterica
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-10-31 Classification: HYDROLASE |
 |
Organism: Salmonella typhimurium (strain lt2 / sgsc1412 / atcc 700720)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-10-24 Classification: HYDROLASE Ligands: ASP |
 |
N-Terminal Domain Of Fact Complex Subunit Spt16 From Eremothecium Gossypii (Ashbya Gossypii)
Organism: Ashbya gossypii (strain atcc 10895 / cbs 109.51 / fgsc 9923 / nrrl y-1056)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-08-15 Classification: DNA BINDING PROTEIN |

