Search Count: 31
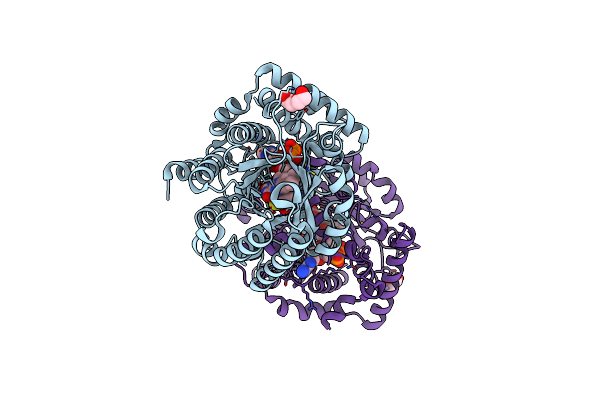 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Prop-2-Ynylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X79 |
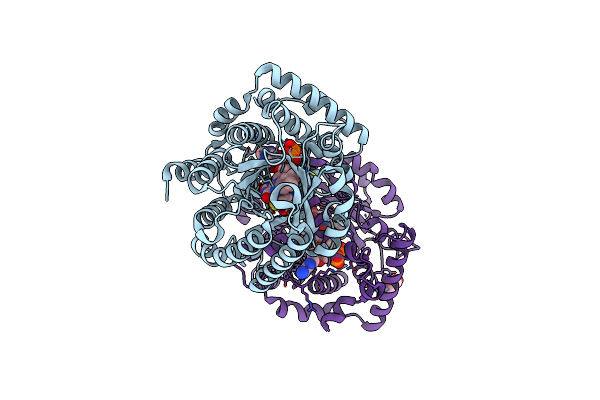 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With 2-(Cyanomethylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, X7K, PEG |
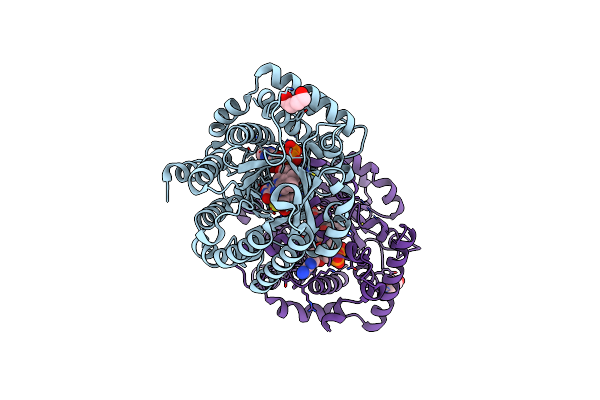 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Complexed With (Allylthio)Acetic Acid
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, X7Q |
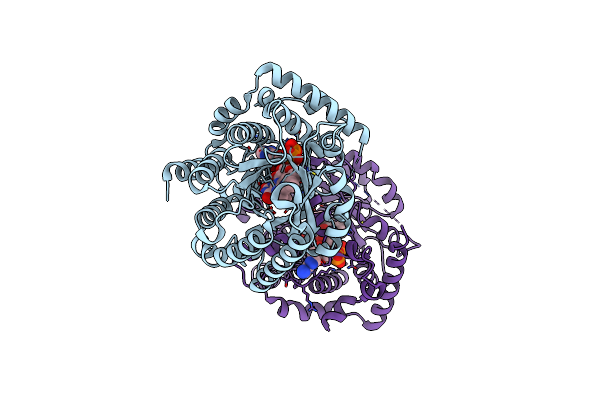 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine(Replicate #1)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) With The Fad N5 Modified With Propanal Resulting From Inactivation With N-Allylglycine (Replicate #2)
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, FAD, CBG, PEG |
 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By Propanal Resulting From Inactivation With N-Allylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: FDA, CBG, NAD, MG, SO4, PEG, A1AV8, PGE |
 |
Structure Of Proline Utilization A With 1,3-Dithiolane-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJD, FAD, NAD, SO4, MG, PGE, PEG, PG4 |
 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By 1,3-Dithiolane
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJJ, MG, NAD, SO4, PGE, PEG, PG4 |
 |
Structure Of Proline Utilization A With Tetrahydrothiophene-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJM, UJP, PGE, FAD, NAD, SO4, MG, PEG |
 |
Structure Of Proline Utilization A With The Fad Covalently Modified By Tetrahydrothiophene
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJG, NAI, SO4, PGE, FMT, MG, PEG |
 |
Structure Of A Stable Interstrand Dna Crosslink Involving An Da Amino Group And An Abasic Site
|
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-09-09 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Structure Of Proline Utilization A With The Fad Covalently Modified By L-Thiazolidine-2-Carboxylate And Three Cysteines (Cys46, Cys470, Cys638) Modified To S,S-(2-Hydroxyethyl)Thiocysteine
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: FDA, T2C, NAD, PEG, SO4, FMT |
 |
Structure Of Proline Utilization A With The Fad Covalently Modified By L-Thiazolidine-2-Carboxylate
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2020-03-18 Classification: OXIDOREDUCTASE Ligands: FAD, NAD, MG, T2C, PEG, SO4, PGE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE Ligands: UN1, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE Ligands: EDO, NAD, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE Ligands: EDO, CL, NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-11-06 Classification: OXIDOREDUCTASE Ligands: NAD |

