Search Count: 13
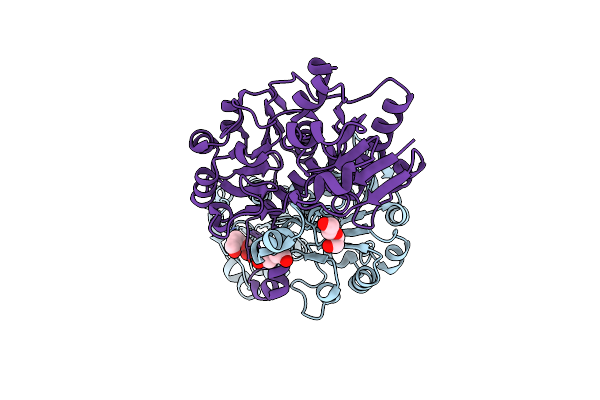 |
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: PEG, PGE |
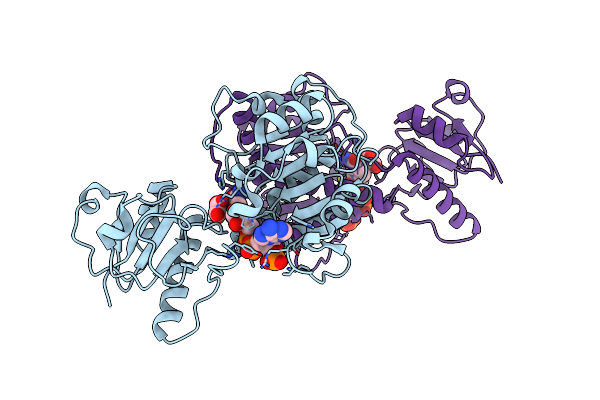 |
Crystal Structure Of Nadph-Dependent 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadph And Oxalate
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: NDP, OXD |
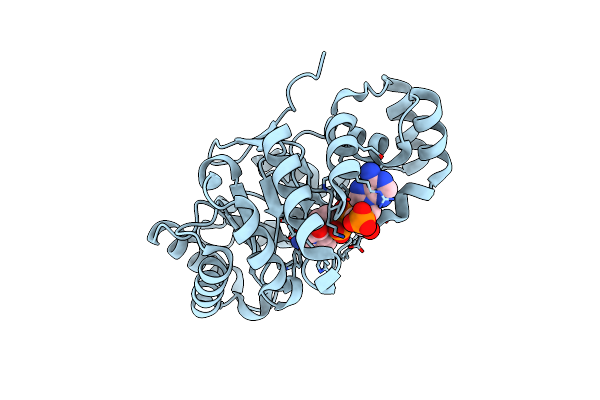 |
Crystal Structure Of Aldo-Keto Reductase From Sinorhizobium Meliloti 1021 In Complex With Nadph
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: NDP |
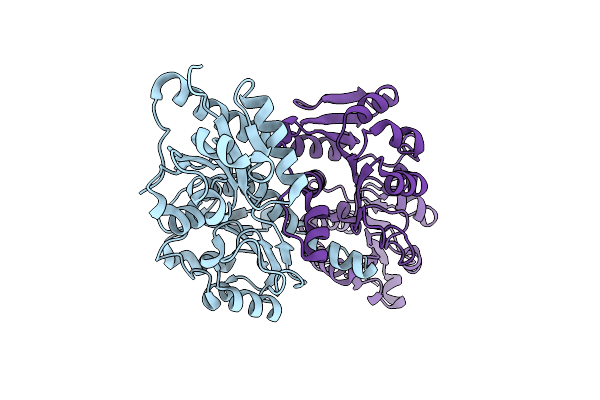 |
Crystal Structure Of Nadp-Dependent Dehydrogenase From Rhodobacter Sphaeroides
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-07-01 Classification: OXIDOREDUCTASE |
 |
Probable 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadp, Hepes And L(+)-Tartaric Acid
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-06-17 Classification: OXIDOREDUCTASE Ligands: EPE, PEG, NAP, TLA, NA, CL |
 |
Crystal Structure Of Sam-Dependent Methyltransferase From Geobacter Sulfurreducens In Complex With S-Adenosyl-L-Homocysteine
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: SAH, CL |
 |
Crystal Structure Of Sam-Dependent Methyltransferase From Bacteroides Fragilis In Complex With S-Adenosyl-L-Homocysteine
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: ETE, PG4, SAH |
 |
Crystal Structure Of Nadp-Dependent Dehydrogenase From Rhodobactersphaeroides In Complex With Nadp And Sulfate
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-05-20 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, PGE, GOL, PEG |
 |
Organism: Polaromonas sp. js666
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-21 Classification: OXIDOREDUCTASE Ligands: CL, NA |
 |
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Xanthobacter Autotrophicus Py2
Organism: Xanthobacter autotrophicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-01-14 Classification: OXIDOREDUCTASE |
 |
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2014-12-31 Classification: OXIDOREDUCTASE |
 |
Probable 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadph
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-12-31 Classification: OXIDOREDUCTASE Ligands: NDP, CL, PEG |
 |
Crystal Structure Of A Putative Oxidoreductase From Sinorhizobium Meliloti 1021 In Complex With Nadp
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-06-11 Classification: OXIDOREDUCTASE Ligands: NAP, GOL |

