Search Count: 11
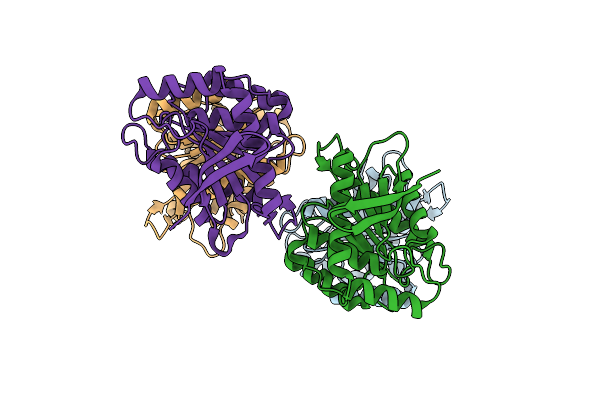 |
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-05-14 Classification: HYDROLASE |
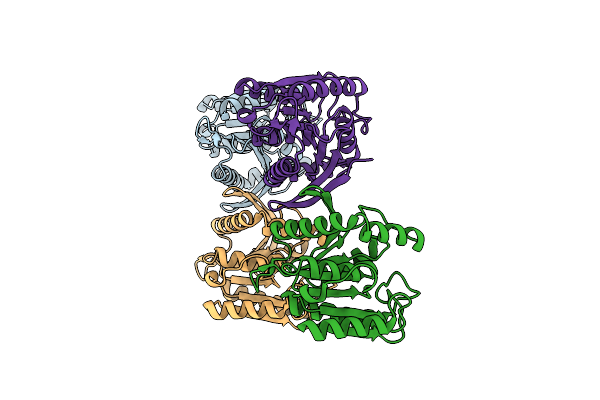 |
Crystal Structure Of Thermostable Dienelactone Hydrolase. Monoclinic Space Group.
Organism: Hydrogenobacter thermophilus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2025-05-14 Classification: HYDROLASE |
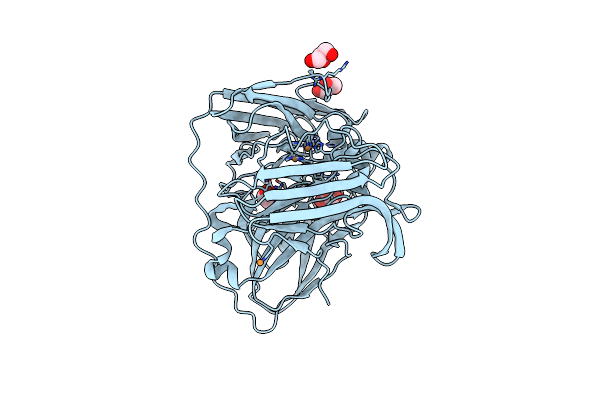 |
Thermus Thermophilus Hb27 Laccase (Tth-Lac) Mutant With Partial Deletion Of Beta-Hairpin Sequence
Organism: Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-02-05 Classification: METAL BINDING PROTEIN Ligands: CU, GOL |
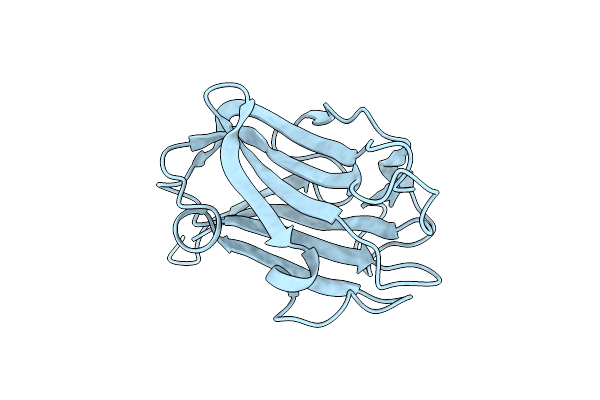 |
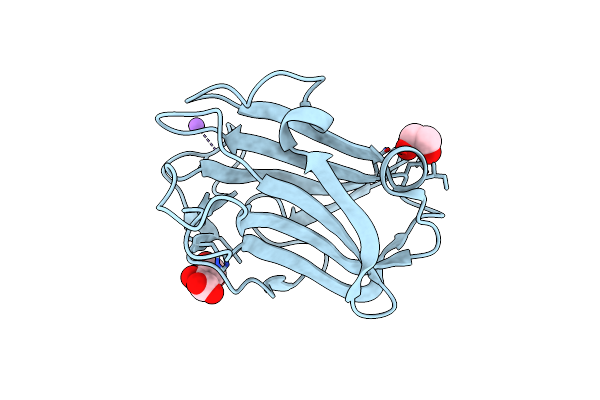
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum Complexed With Iodine Atoms
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: IOD |
 |
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: EDO, NA, LMR |
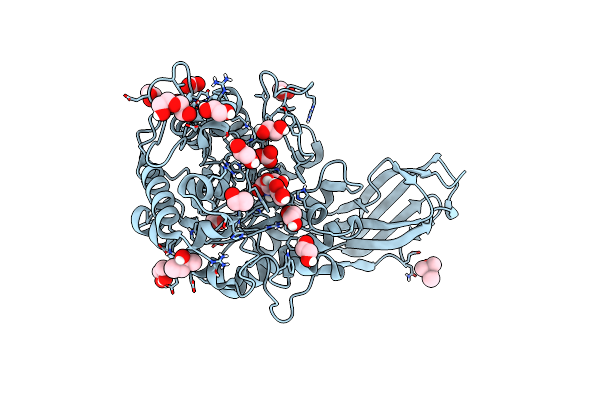 |
Organism: Thermotoga petrophila rku-1
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: MPD, EDO, ACT, CO3, GOL, PEG, TRS, CL, NA |
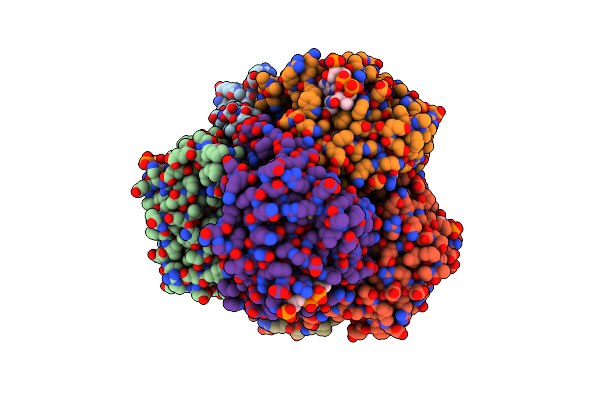 |
Structure Of An Enoyl-Coa Hydratase/Aldolase Isolated From A Lignin-Degrading Consortium
Organism: Uncultured organism
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-08 Classification: LYASE Ligands: COA, B3P |
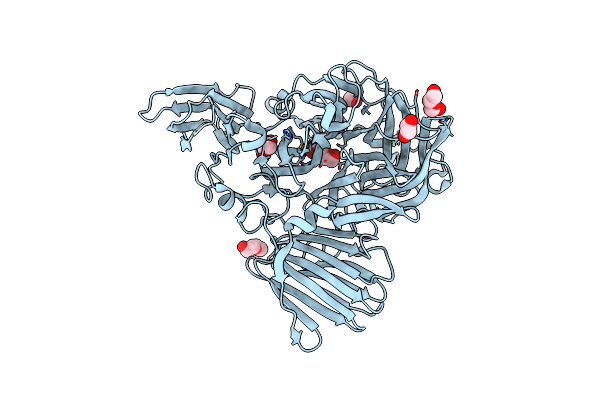 |
Structure Of Glycoside Hydrolase Family 32 From Bifidobacterium Adolescentis
Organism: Bifidobacterium adolescentis (strain atcc 15703 / dsm 20083 / nctc 11814 / e194a)
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2020-01-22 Classification: HYDROLASE Ligands: EDO, PEG |
 |
Organism: Bacillus subtilis
Method: SOLUTION NMR Release Date: 2015-03-25 Classification: CELL CYCLE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2015-03-18 Classification: CELL CYCLE Ligands: PO4 |
 |
Organism: Xylella fastidiosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-03-02 Classification: TRANSFERASE Ligands: CL, GSH |

