Search Count: 43
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: ZN, 3PE, DXC, PLP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: ZN, 3PE, DXC, XBE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2024-06-26 Classification: MEMBRANE PROTEIN Ligands: ZN, 3PE, DXC, HCZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-10-20 Classification: TRANSFERASE Ligands: V0K |
 |
Organism: Bacillus anthracis, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: TOXIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2019-10-09 Classification: CYTOKINE |
 |
Spectroscopic And Structural Study Of Qw, A Egfp Mutant Showing Photoswitching Properties
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-02-06 Classification: FLUORESCENT PROTEIN |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: CL |
 |
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.80 Å Release Date: 2019-01-23 Classification: HYDROLASE Ligands: CL |
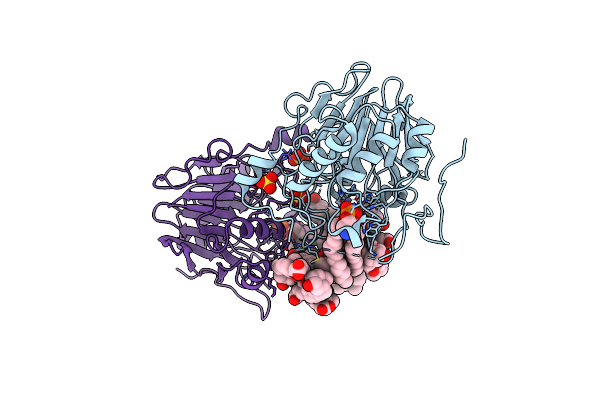 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-06-17 Classification: HYDROLASE Ligands: ZN, 3PE, DXC, SO4 |
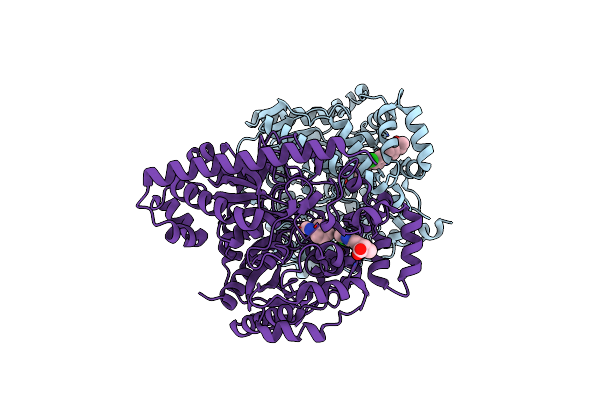 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-01-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CL, OHO, 0LA |
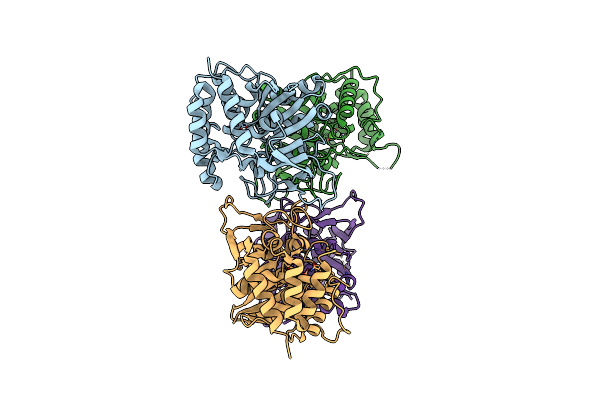 |
Crystal Structure Of The E. Coli Pyrimidine Nucleoside Hydrolase Yeik (Apo-Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-12-01 Classification: HYDROLASE Ligands: CA |
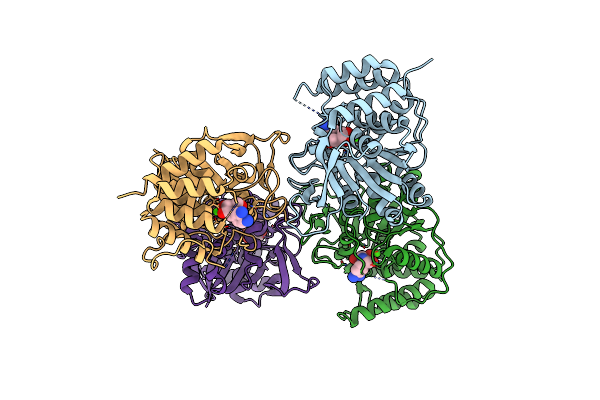 |
Crystal Structure Of The E. Coli Pyrimidine Nucleosidase Yeik Bound To A Competitive Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-12-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA, DNB |
 |
Crystal Structure Of The E.Coli Riha Pyrimidine Nucleosidase Bound To A Iminoribitol-Based Inhibitor
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-02-09 Classification: HYDROLASE Ligands: BME, CA, DNB |
 |
Structural Basis For The Broad-Spectrum Inhibition Of Metallo-{Beta}-Lactamases: L1- Is38 Complex
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-08-28 Classification: HYDROLASE Ligands: ZN, SO4, I38 |
 |
Crystal Structure Of The Zinc Carbapenemase Cpha In Complex With The Inhibitor D-Captopril
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2007-07-17 Classification: HYDROLASE Ligands: ZN, SO4, MCO, GOL |
 |
Crystal Structure Of The Zinc-Beta-Lactamase L1 From Stenotrophomonas Maltophilia (Inhibitor 3)
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-05-29 Classification: HYDROLASE Ligands: ZN, SO4, L13 |
 |
Spectroscopic And Structural Study Of The Heterotropic Linkage Between Halide And Proton Ion Binding To Gfp Proteins- E2(Gfp) Apo Form
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2007-05-15 Classification: LUMINESCENT PROTEIN |
 |
Spectroscopic And Structural Study Of The Heterotropic Linkage Between Halide And Proton Ion Binding To Gfp Proteins: E2(Gfp)-Cl Complex
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2007-05-15 Classification: LUMINESCENT PROTEIN Ligands: CL |
 |
Spectroscopic And Structural Study Of The Heterotropic Linkage Between Halide And Proton Ion Binding To Gfp Proteins: E2(Gfp)-Br Complex
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-05-15 Classification: LUMINESCENT PROTEIN Ligands: BR |

