Search Count: 1,366
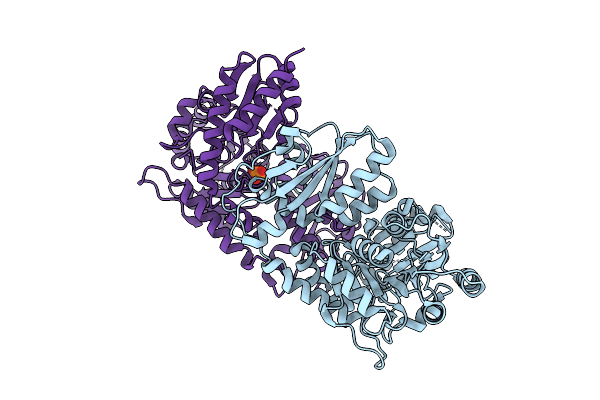 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LYASE Ligands: PO4, CL |
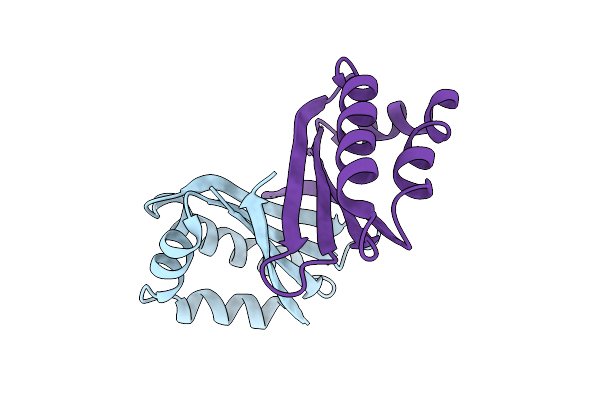 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
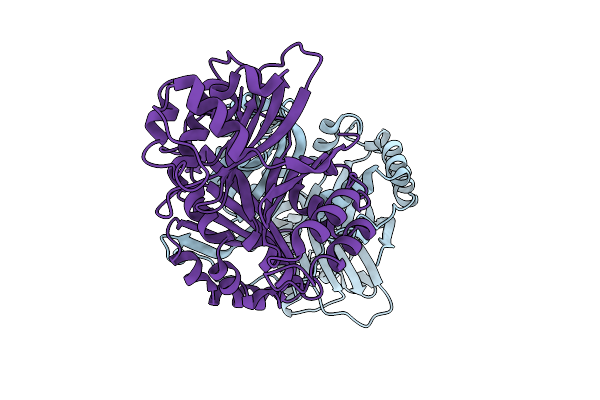 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
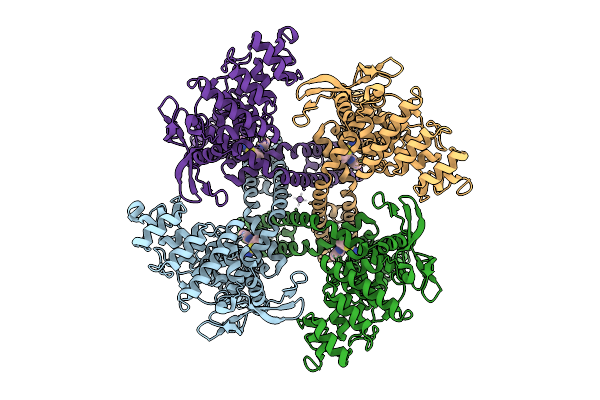 |
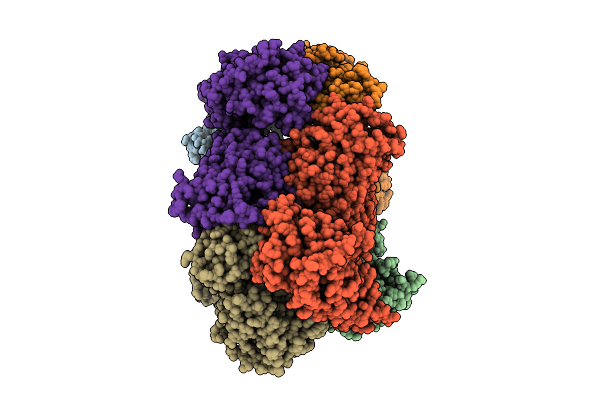
Organism: Escherichia coli k-12, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1D6R, NA |
 |
Organism: Escherichia coli k-12, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1D6W, NA |
 |
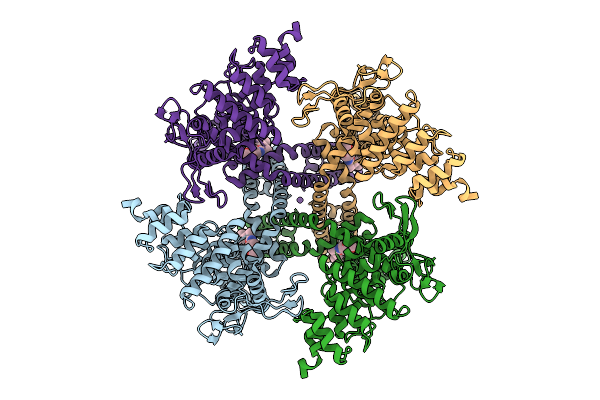
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ANTIBIOTIC |
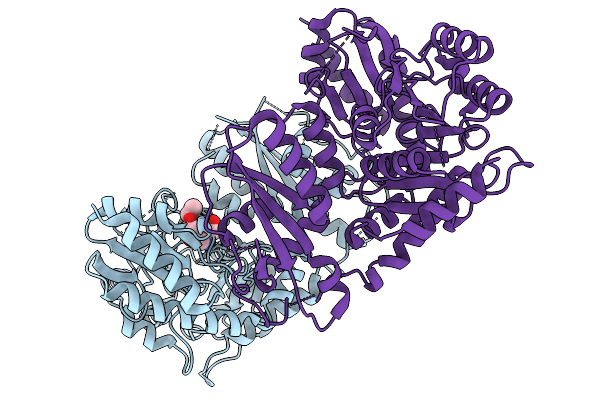 |
Gamma-Lyase Cndf In Complex With Pyridoxal 5'-Phosphate, L-Homoserine And Ethyl Acetoacetate
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: HSE, EAC |
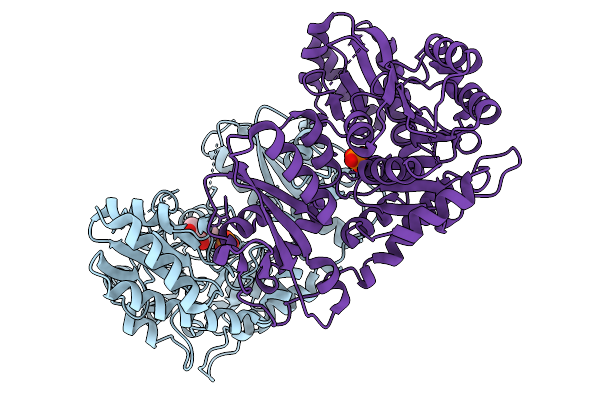 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: PO4, EAC |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: OT5 |
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Coupling Of Polymerase-Nucleoprotein-Rna In An Influenza Virus Mini Ribonucleoprotein Complex
Organism: Influenza a virus (a/victoria/3/1975(h3n2))
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN/RNA |
 |
Organism: Epilithonimonas lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE Ligands: C2E, CA |
 |
Organism: Epilithonimonas lactis
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: HYDROLASE Ligands: C2E |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Spodoptera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: AND |

