Search Count: 470
 |
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Synthetic construct, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: A1EKK |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: A1EKK |
 |
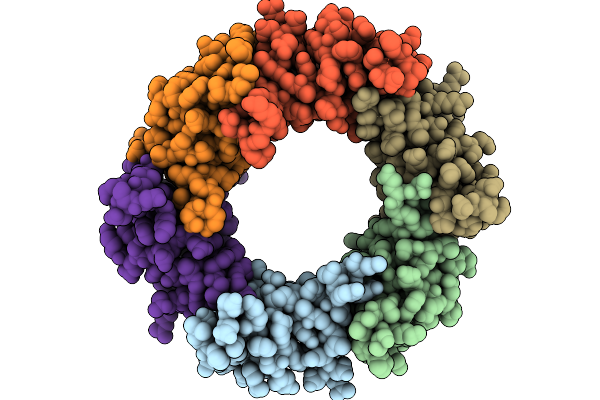
Structure Of The Bacteroides Fragilis Nctc9343 T6Ss Hcp2-Hcp3 Heterohexamer In Complex With The Effector Bte1
Organism: Bacteroides fragilis nctc 9343
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacteroides fragilis nctc 9343
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN |
 |
Organism: Pectobacterium atrosepticum scri1043
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
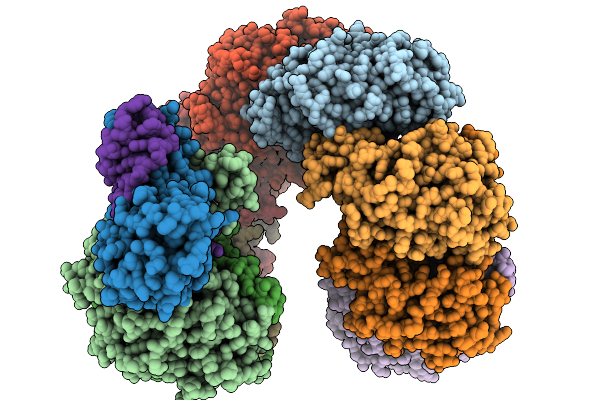
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens dsm 198
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
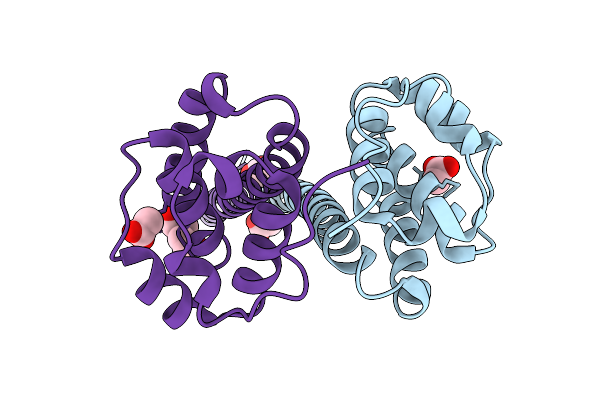
Crystal Structure Of A Transaminase Pata From Pseudonocardia Ammonioxydans In Complex With Plp And Llp
Organism: Pseudonocardia ammonioxydans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PLP |
 |
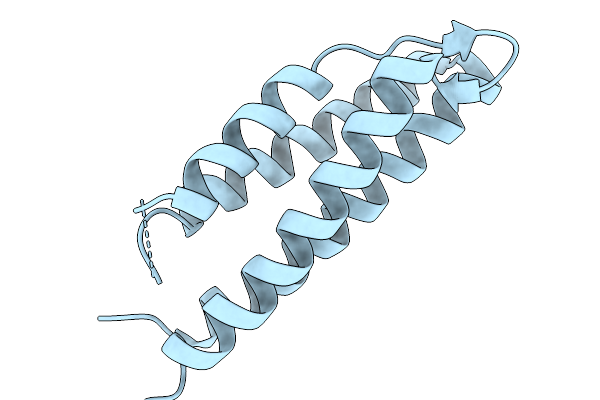
Outer Membrane Lipoprotein Qseg Of Salmonella Enterica Serovar Typhimurium Sl1344
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-11-19 Classification: SIGNALING PROTEIN Ligands: ACT, PEG |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
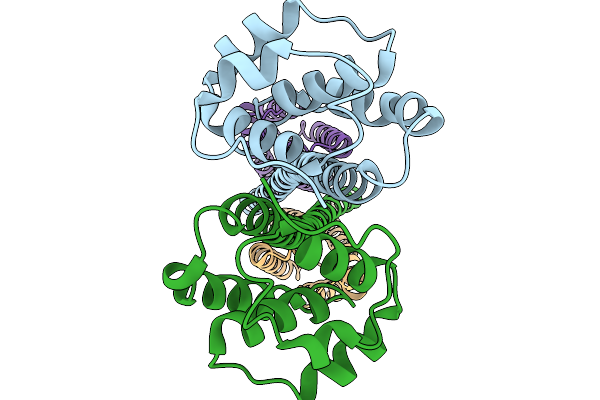 |
Structure Of Outer Membrane Lipoprotein Qseg And Histidine Kinase Qsee Complex
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
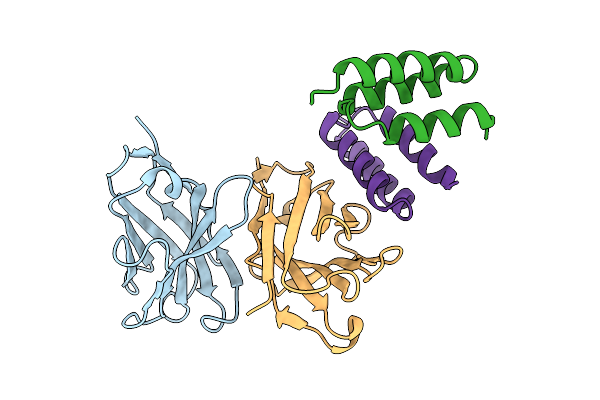 |
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
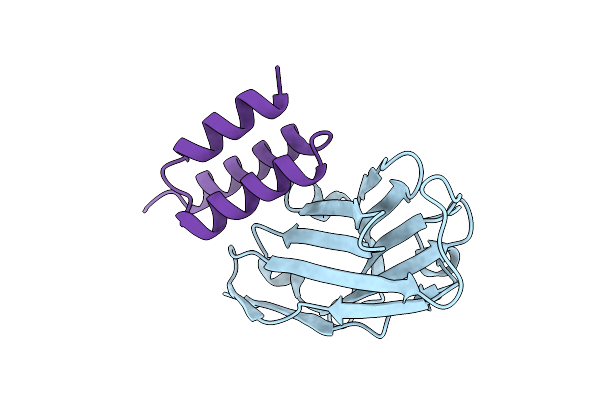 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
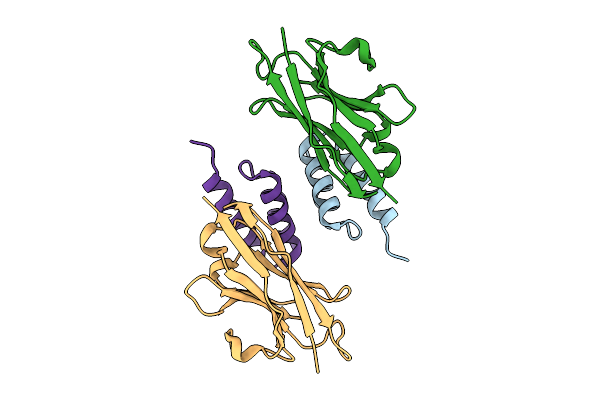 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Resolution:2.32 Å Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Resolution:2.94 Å Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Resolution:3.05 Å Release Date: 2025-10-29 Classification: TOXIN |

